
Metastasis-associated gene 1 (MTA1) enhances cisplatin resistance of malignant pleural mesothelioma by ATR-Chk1-mediated DNA repair
Introduction
Malignant pleural mesothelioma (MPM) is a rare malignant cancer with a poor prognosis and few treatment options (1). More than 95% patients are diagnosed at a later stage and the median overall survival time is less than 9 months (2). The primary cause of short overall survival is insensitivity to platinum-based chemotherapeutics, which are still the main treatment for MPM (3). Therefore, understanding the molecular mechanism of chemotherapy resistance in MPM remains a challenge for clinicians and scientists.
Multiple mechanisms are involved in the anti-cancer effects of cisplatin, yet the most important mechanism is to cause DNA replication disorder, thereby inhibiting the mitosis and proliferation of cancer cells (4). Although cisplatin treatment is initially effective, cells quickly develop chemotherapy resistance, leading to treatment failure (5). Nevertheless, DNA lesions induced by cisplatin can be repaired by multiple pathways, of which the 2 most important are the nucleotide excision repair and the mismatch repair systems (6,7). Once the systems are activated, ataxia telangiectasia mutated (ATM) and ATM and rad3 related (ATR) will be phosphorylated, thereby blocking cells in the S and G2 phases and re-establishing DNA integrity (8). ATM is mainly activated by the ionizing radiation-induced DNA damage response. As the downstream of ATM, ATR is not only activated by ATM but also activated by chemotherapeutic agents, such as cytotoxic agents and antimetabolites (9,10). When activated by DNA lesions, ATR phosphorylates its most prominent substrate and downstream effector, checkpoint kinase 1 (Chk1), which protects tumor genomes against damage caused by chemotherapeutic agents (11). The ATR-Chk1 pathway then phosphorylates multiple substates, including DNA replication, DNA repair, and cell cycle proteins, which interrelates chemoresistance of tumor cells (12). For instance, Chk1 inactivates the cyclin-dependent kinase/cyclin complex through M phase inducer phosphatase Cdc25 (Cdc25), rapidly halting DNA replication in the S phase and preventing cells with damaged DNA from entering mitosis (13). It has been well established that the ATR-Chk1 pathway promotes homologous recombination by regulating the phosphorylation of RAD51 recombinase (RAD51), FA complementation group E (FANCE), and FA complementation group D2(FANCD2), which is a major pathway required for the repair of DNA double-strand breaks and collapsed replication forks (14-16). Therefore, the inhibition or knockdown of ATR and/or Chk1 expression can increase the sensitivity of tumor cells to a mass of DNA-damaging agents, especially DNA cross-linking agents (cisplatin) (17).
MTA1, an important part of nucleosome remodeling and histone deacetylation complexes, is widely overexpressed in multiple cancers and participates in tumorigenesis, metastasis, and chemoresistance (18,19). In our previous study, we found that MTA1 plays an oncogenic role in promoting the metastasis of MPM via E-cadherin, leading to the poor prognosis of patients (20). Interestingly, a previous study revealed MTA1 takes part in the ATR-Chk1-mediated DNA damage checkpoint pathway, which reduces the sensitivity of tumor cells to Ultraviolet (UV)-induced DNA damage (21). According to the mechanism of the ATR-Chk1 pathway which participates in chemoresistance and the association of ATR and MTA1, we hypothesized that MTA1 may also play an important role in the chemoresistance and cell cycle regulation of MPM cells. In recent study, we observed that cisplatin promotes the expression of the MTA1 protein by inhibiting its ubiquitination. However, MTA1 enhances cisplatin resistance of MPM cells by enhancing G2/M cell cycle delay. Furthermore, MTA1 regulates the phosphorylation of the ATR-Chk1 pathway, protecting tumor genomes against damage caused by chemotherapeutic agents. Therefore, our findings indicate that MTA1 enhances the chemoresistance of MPM cells by regulating the ATR-Chk1 pathway. We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/atm-21-941).
Methods
Patients and tissue samples
Tissue samples used in the present study were obtained from 39 MPM patients with new cases. The information of patients was collected in our previous study (20). The study was approved by the Ethical Committee of the First Affiliated Hospital of Soochow University (Suzhou, China), and written informed consent was obtained from each patient. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Western blotting and immunoprecipitation
Cell protein was extacted and quantitated by protein assay (Beyotime, Shanghai, China). Proteins were fractionated by sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a polyvinylidene fluoride membrane, blocked in 4% dry milk at room temperature for 1 h, and immunostained with primary antibodies at 4 °C overnight using anti-MTA1 (1:2,000; Abcam, Cambridge, MA, USA), anti-ATR (1:1,000; Abcam, USA), anti-p-ATR (Ser428, 1:1,000; Abcam, USA), anti-ATR-interacting protein (ATRIP) (1:1,000; Abcam, USA), anti-Chk1 (1:10,000; Abcam, USA), anti-p-Chk1(1 µg/mL; Abcam, USA), anti- RAD17 checkpoint clamp loader component (RAD17) (1:500; Abcam, USA), anti- chromatin-binding protein RAD9 (RAD9) (1:2,000; Abcam, USA), anti- DNA topoisomerase II binding protein 1 (TopBP1) (1:1,000; Abcam, USA), anti-Claspin (1:500; Abcam, USA), and anti- glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:1,000; KangChen, Beijing, China). The bands were detected via a chemiluminescent detection system (Thermo Fisher Scientific, Rockford, IL, USA) and exposed in the Molecular Imager ChemiDoc XRS System (Bio-Rad, Hercules, CA, USA).
For the immunoprecipitation (IP) analysis, 30 µL of protein A/G PLUS-agarose (Santa Cruz Biotechnology, Dallas, TX, USA) and 1 µg of antibody were added to 1–2 mg of protein extracts. The immunoprecipitates were collected by centrifugation in a microcentrifuge at 14,000 g for 5 min, and the supernatant was discarded after overnight incubation at 4 °C on a rocker platform. Nonidet P-40 buffer (50 mM Tris-HCl, pH 8.0, 0.5% Nonidet P-40, 10% glycerol, 150 mM NaCl, 2 mM MgCl2, and 1 mM ethylenediaminetetraacetic acid with protease inhibitors) washed the pellet 5 times and then dissolved in a sample buffer for polyacrylamide gel electrophoresis (SDS-PAGE).
Lentivirus production and transduction
MPM cells which were transfected by lentivirus containing pLL3.7-empty and pLL3.7-shMTA1 were obtained from D. Yun Chen from Nanjing Medical University. The uppercase letters represent MTA1-specific sequence, and lowercase letters represent hairpin sequences. Recombinant lentivirus was generated from 293 T cells using calcium phosphate precipitation. MSTO-211H and H2452 were transfected with lentivirus using polybrene (8 µg/mL).
Cell culture and establishment of cisplatin-resistant MPM cell lines
MSTO-211H, NCI-H2452, and 293 T cell lines [American Type Culture Collection (ATCC), Manassas, VA, USA] were employed for this study. MSTO-211H and H2452 were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA), while 293 T cells were cultured in Dulbecco’s modified Eagle’s medium and high glucose medium supplemented with 10% fetal bovine serum. To establish cisplatin-resistant subclones, MSTO-211H and NCI-H2452 cells were cultured with step-wise increasing concentrations of cisplatin [concentrations from the half-maximal inhibitory concentration (IC50) to 10 folds of IC50 of wildtype cells] for 15 months, and 2 cell cisplatin-resistant lines, MSTO- DDP and H2452-DDP, were established. All cells were cultured in a humidified 37 °C incubator with 5% CO2.
Cell counting Kit-8 (CCK-8) assay
Cells were plated at a density of 30,000 cells per well in a 96-well plate at 37 °C with 5% CO2 for 24 h. Then, 10% CCK-8 reagent was added to each well. The optical density at 450 nm was measured on a microplate reader after incubation for 4 h (Labsystems, Helsinki, Finland).
Immunohistochemical staining
Tissues which were fixed in 4% paraformaldehyde and cut from a paraffin block to 5-µm thickness were dewaxed with xylene and rehydration with a graded series of ethanol. The slides were heated in the autoclave for 3 min using citrate buffer (pH 6.0) and incubated with primary antibody MTA1 (1:1,000; Abcam, USA) at 4 °C overnight. Blocking serum or antibody dilution buffer was prepared as the negative control. The primary antibodies utilized were all the same as for the Western blot analysis. Photographs were taken by microscope (ECLIPSE 50i; Nikon, Tokyo, Japan) and NIS-Elements version 4.0 (Nikon, Tokyo, Japan). Average values of integrated optical density were obtained from 5 random fields per slide by using Image-Pro Plus software (version 5.0) (Media Cybernetics, Maryland, USA). Every data was acquired from detections repeated 3 times at least.
Real-time polymerase chain reaction (PCR) analysis
The mRNA levels of MTA1 were detected by real-time PCR using SYBR Premix Ex Taq (TAKARA, Tokyo, Japan). The expression of β-actin was prepared as the negative control. Amplification conditions, primers, and probes sequences for MTA1 and β-actin were from a previous study (20). All procedures were done in accordance with the MIQE guidelines (22).
Crystal violet assay
Cells in a 6-well plate were incubated for 24 h at 37 °C. The medium was aspirated and the cells were washed twice in a gentle stream of tap water. The water from the wells was immediately aspirated. A total of 1 mL of 0.5% crystal violet staining solution was added to each well, and the wells were incubated for 20 min at room temperature on a bench rocker with a frequency of 20 oscillations per minute. The plates were washed 4 times and the plate was inverted on filter paper. The plate was tapped gently for 2 h to remove any remaining liquid. A total of 4 mL of methanol was added to each well, and the plate with its lid on was incubated for 20 min at room temperature on a bench rocker with a frequency of 20 oscillations per minute.
Cell cycle analysis
Cells were trypsinized and then centrifuged. The pellet was washed 2–3 times with 1× phosphate buffer saline (PBS) to exclude the remaining culture medium. The cells were fixed with 90% methanol to −20 °C for 10 min. Afterwards, the cells were washed twice with 1× PBS and then incubated in a solution containing 50 µg/mL of propidium iodide and 100 µg/mL of RNase A for 30 min at 37 °C in the dark. After incubation, the cells were centrifuged and the pellet was resuspended in 1–1.5 mL of 1× Focusing Fluid (Thermo Fisher Scientific Inc., USA). Flow cytometry analysis was conducted using the Attune NxT Acoustic Focusing Cytometer (Thermo Fisher Scientific Inc., USA); in each run, at least 10,000 events were collected to 200 µL/min speed, using the channel BL2-A (574/26 nm filter). The histograms, dot plots, and density plots were made using Attune NxT SW version 1.1 software. The percentage of fibroblasts in the different phases of cells cycle (G0/G1, S, and G2/M) was estimated.
Phosphohistone H3 staining and assay
Cells were harvested at the indicated time points by trypsinization and fixed with 70% ethanol. The cell pellet was suspended in 100 µL of PBS containing 0.5% bovine serum albumin and 1:50 diluted phosphohistone H3 (Ser10) antibody, followed by staining with fluorescein isothiocyanate-labeled secondary antibody. The cells were stained with 50 µg/mL propidium iodide and 10 µg/mL RNase A after 1 h of incubation at room temperature. Cellular fluorescence was determined by a BD Biosciences FACScan (BD Biosciences, San Jose, USA), and the data were analyzed using Cell Quest software (Becton Dickinson, San Jose, USA).
Statistical analysis
Statistical analysis was performed using GraphPad Prism (version 5.01; GraphPad Software, La Jolla, CA, USA) statistical software. The Student’s t-test and paired t-test were used to analyze significance between independent groups and paired materials, respectively. The χ2-test was used to test the significance of observed differences in proportions, except when the cell size was less than 5groups (Fisher’s exact tests). P<0.05 indicated statistical significance.
Results
MTA1 protein was induced following exposure to cisplatin in MPM
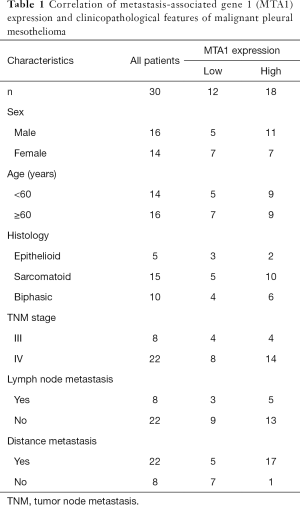
To explore the clinical relevance of MTA1 expression in the regulation of cisplatin resistance in MPM, we investigated 9 MPM patients with different pathological types who developed resistance to cisplatin. We found that MTA1 protein level increased significantly after cisplatin resistance in the patients (Figure 1A,B). However, there was no change in the MTA1 mRNA level (Figure 1C). Subsequently, to further confirm the effect of cisplatin on the MTA1 protein, immunohistochemistry was used to detect tissues from 2 patients, and the results suggested that the expression of the MTA1 protein was strongly positive after cisplatin resistance (Figure 1D). Thirty MPM patients were divided into 2 groups using a semiquantitative immunoreactivity scoring system, as reported in our previous study (Table 1) (20). Our findings indicated that after cisplatin-based chemotherapy, MPM patients with a high expression of MTA1 protein had significantly reduced progression-free survival (PFS) time than patients with a low expression of MTA1 protein (Figure 1E). These results suggest that the expression of MTA1 protein can be induced by cisplatin chemotherapy and could be associated with the prognosis of MPM patients.
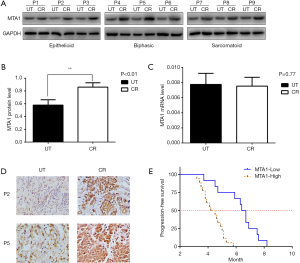
Full table
MTA1 enhanced cisplatin resistance of MPM cells
To further clarify the relationship between MTA1 and cisplatin in vitro, 2 different cell lines (H2452 and MSTO) were employed in our study. We successfully constructed 2 cell lines with a low expression of the MTA1 protein (H2452-shMTA1 and MSTO-shMTA1) by infecting with lentivirus plasmid (pLL3.7), as reported in our previous study (Figure 2A) (20). We then detected the IC50 values of cisplatin in each cell line via CCK-8 assay (Figure 2B,C). H2452 and MSTO showed 1.9- and 1.8-fold increased IC50 values for H2452-shMTA1 and MSTO-shMTA1 cells, respectively (Figure 2D). To further confirm the effect of MTA1 on cisplatin toxicity, we established cisplatin-resistant cell lines (H2452-DDP and MSTO-DDP) and detected MTA1 protein expression. We found that the expression of the MTA1 protein was significantly increased in H2452-DDP and MSTO-DDP compared with H2452 and MSTO (Figure 2E). Meanwhile, we detected the MTA1 protein expression in MPM DDP-resistant cells transfected with siMTA1. As shown in Figure 2F, siMTA1 reduced the level of MTA1 protein expression in MPM DDP-resistant cells. Subsequently, we detected the IC50 values of cisplatin in H2452-DDP and MSTO-DDP transfected with siMTA1 (Figure 2G,H). siMTA1 showed reduced 1.8- and 1.7-fold IC50 values for H2452-DDP and MSTO-DDP cells, respectively (Figure 2I). In clonogenic assays, we found that low MTA1 expression cell lines (H2452-shMTA1 and MSTO-shMTA1) treated with cisplatin caused a marked inhibition of proliferation compared with H2452 and MSTO (Figure 2J,K). Similarly, we also found that cisplatin combined with siMTA1 could remarkably reduce the proliferation of H2452-DDP and MSTO-DDP cells (Figure 2L). We then examined the MTA1 protein levels in H2452-DDP and MSTO-DDP cells, which were treated with cisplatin and siMTA1 (Figure 2M,N). Collectively, these results suggest that MTA1 protein expression is involved in the cisplatin resistance of MPM cells.
Cisplatin stabilized the expression of the MTA1 protein by inhibiting its ubiquitination
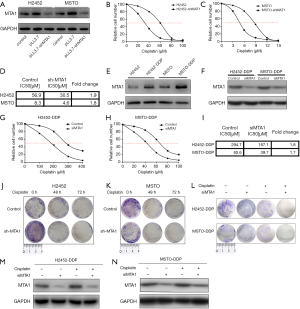
Cisplatin induces the MTA1 protein in MPM patients, whereas the MTA1 mRNA level has no change, indicating that a post-transcriptional mechanism might be involved in the regulation of the MTA1 protein. To verify this, MPM cells were exposed to cisplatin at the indicated time points for Western blotting and quantitative real-time PCR assay. As shown in Figure 3A, MTA1 protein expression gradually increased and reached its peak at 48 h, while the MTA1 mRNA level did not change significantly. On the basis of this observation, we then examined the MTA1 expression at the indicated doses of cisplatin for 48 h and found that MTA1 protein expression increased with cisplatin dose, while the MTA1 mRNA level did not change (Figure 3B). Interestingly, these results were consistent with the clinical data in Figure 1. Subsequently, a eukaryote protein synthesis inhibitor (cycloheximide) was employed to observe the effect on the degradation rate of the endogenous MTA1 protein in MPM cells treated with or without cisplatin. We found that cisplatin significantly stabilized the expression of the MTA1 protein compared with untreated controls (Figure 3C). These findings suggest that cisplatin induces the MTA1 protein through its post-transcriptional regulation.
A previous study demonstrated that UV radiation stabilizes the MTA1 protein by inhibiting its ubiquitination in HaCaT cells (aneuploid immortal keratinocyte cell line) (21). Therefore, we hypothesized that cisplatin might promote MTA1 protein expression in the same way. To verify this, MSTO cells were pretreated with MG-132 (a proteasome inhibitor) to enhance the signal of ubiquitination of endogenous MTA1, as reported in a previous study (21), and then treated with or without cisplatin. The MSTO protein was subjected to IP analysis with an anti-MTA1 antibody and immunoglobulin G control, followed by Western blot analysis with indicated antibodies. The findings indicated that cisplatin could significantly reduce the ubiquitination of endogenous MTA1 (Figure 3D). Taken together, these results indicate that cisplatin promotes MTA1 protein stability, at least in part, by inhibiting ubiquitin.
MTA1 enhanced G2/M cell cycle delay in MPM cells treated with cisplatin
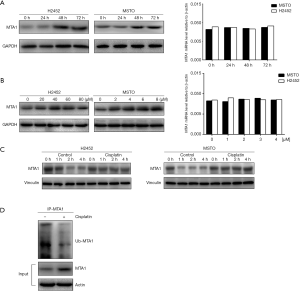
DNA damage induced by chemotherapy can activate the cell cycle checkpoint, which arrests tumor cells in the S or G2/M phase for repair and prevent tumor cells with DNA damage from entering mitosis (23). We next sought to detect whether MTA1 expression affects the cell cycle of MPM cells treated with cisplatin via flow cytometric analysis. As shown in Figure 4A, cisplatin successfully arrested the H2452 cells in the G2/M phase. Nevertheless, knocking down the expression of MTA1 in H2452 cells attenuated this effect of cisplatin. The average value of the G2/M cell cycle from 3 independent experiments was analyzed (Figure 4B). These results were also confirmed in MSTO cells (Figure 4C,D). We then detected the cell cycle of MPM DDP-resistant cells treated with or without cisplatin and siMTA1. We found that the percentage of the G2/M cell cycle did not change significantly when MPM DDP-resistant cells were treated with cisplatin or siMTA1 alone. However, once the cells were exposed to cisplatin combined with siMTA1, the percentage of G2/M cell cycle was significantly reduced (Figure 4E,F,G,H).
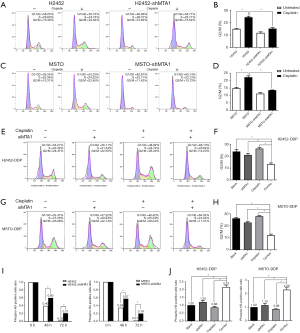
A previous study demonstrated that Phosphohistone-H3-positive cells could distinguish mitotic cells to G2 cells (24). Therefore, we detected the percentage of Phosphohistone-H3-positive cells in cisplatin-treated MPM cells by flow cytometric analysis. As shown in Figure 4I, JenyH2452-shMTA1 and MSTO-shMTA1 cells presented a higher percentage of Phosphohistone-H3-positive cells compared with normal MPM cells at the corresponding time points after cisplatin treatment. Interestingly, the percentage of Phosphohistone-H3-positive cells increased significantly in MPM DDP-resistant cells when exposed to cisplatin combined with siMTA1 (Figure 4J). This result indicated that reducing the level of MTA1 expression can promote cisplatin-treated cells into the mitotic phase. In brief, these results suggest that MTA1 is responsible, at least in part, for G2/M cell cycle delay in MPM cells with cisplatin treatment.
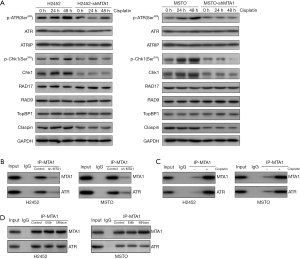
MTA1 regulated and protected tumor genomes from chemotherapeutic drugs via participating in the phosphorylation of the ATR-Chk1 pathway through protein-to-protein interaction
To further analyze the mechanism of MTA1 participating in G2/M cell cycle delay, we detected the related protein expression of the ATR-Chk1 pathway in MPM cells at different time points after cisplatin treatment by Western blotting. As shown in Figure 5A, compared with H2452 and MSTO cells, the expressions of p-ATR, p-Chk1, Chk1, and the Claspin protein were significantly reduced in H2452-shMTA1 and MSTO-shMTA1 cells. The expressions of ATR, ATRIP, RAD17, RAD9, and TopBP1 did not change significantly. As shown in Figure 5B,C, the interaction between MTA1 and ATR was confirmed, and the upregulation of MTA1 and ATR in MPM cells treated with cisplatin was able to be detected via IP assay in H2452 and MSTO cells, respectively. On this basis, to explore whether MTA1 regulation of ATR depends on DNA-protein interactions, we then performed the IP analysis using anti-ATR antibodies in the presence of the DNA intercalator ethidium bromide (EtBR) and Micrococcal nuclease. EtBR disrupts protein-DNA interactions and Micrococcal nuclease digests single-stranded nucleic acids and cleaves double-stranded DNA or RNA (21). The results of the Western blot analysis indicated that Micrococcal nuclease and EtBR treatment had no effect on the binding of ATR to MTA1 (Figure 5D), further indicating that the interaction between MTA1 and ATR is independent on DNA. These results suggest that MTA1 can affect the phosphorylation of the ATR-Chk1 pathway through protein interactions, thereby reducing the negative impact of cisplatin on the genome of MPM cells.
Discussion
MPM, which originates from serosa tissue mesothelial cells (25), is an aggressive malignant tumor with a low survival rate (26,27). The first-line chemotherapy regimen of MPM based on cisplatin plus pemetrexed can extend the median overall survival of MPM patients to 16.1 months (28). One of the main reasons for tumor progression after chemotherapy is the resistance of tumor cells to cisplatin (29). Therefore, to elucidate the mechanisms of resistance of MPM cells to cisplatin can benefit the treatment outcome of patients.
In the present study, we demonstrated that the MTA1 protein was markedly upregulated in MPM specimens of cisplatin-resistant patients, and can affect the PFS of patients in different ways. In addition, we demonstrated a vital role of MTA1 in the process of enhancing cisplatin resistance of MPM cells. Of importance, we further confirmed that MTA1 can protect tumor genomes from cisplatin by affecting the ATR-Chk1 pathway and inducing G2/M cell cycle delay in MPM cells treated with cisplatin. Additionally, cisplatin can also stabilize the expression of the MTA1 protein via inhibiting its ubiquitination. On the basis of these findings, we speculated that MTA1 potentially plays an carcinogenic role in the chemotherapeutic insensitivity of MPM.
MTA1 has been supposed to be closely correlated with carcinogenesis and the progression of various types of human cancers, such as breast cancer (30), gastrointestinal cancer (31-33), non-small cell lung cancer (34), and ovarian cancer (35). Our previous study also showed that MTA1 could promote the metastasis by direct regulation of E-cadherin in MPM patients (20). In addition, increasing evidence has shown that MTA1 overexpression can induce cisplatin resistance in various tumor types, including cervical cancer and nasopharyngeal carcinoma (36,37). Therefore, we conducted further research on the role of MTA1 in MPM. Consistent with the above research, we found that the MTA1 protein was upregulated in cisplatin-resistant MPM cells and was correlated with cisplatin resistance. Of importance, we reported that MPM patients who received cisplatin chemotherapy with a high expression of MTA1 had significantly shorter PFS.
It is well known that DNA damage response (DDR) is critical to protect against genomic instability, and ATR kinase has key roles in DDR (17,38). When cancer cells are treated with radiation therapy or chemotherapeutic drugs, such as cisplatin, the cell cycle checkpoint protein, ATR, and its key downstream regulator, Chk1, halt their pace into mitosis, leading to treatment failure and tumor progression (39). Therefore, a series of agents targeting the ATR-Chk1 pathway have been designed to achieve more ideal chemotherapeutic effectiveness (40-42). Li et al. demonstrated that MTA1 participates in the UV-induced DNA damage checkpoint pathway, which is mediated by the ATR-Chk1 pathway (21), indicating the potential relationship between MTA1 and the ATR-Chk1 pathway. In the present study, we verified the interaction between MTA1 and the ATR-Chk1 pathway in MPM cells treated with cisplatin.
In summary, we demonstrate a vital role for MTA1 in chemotherapy resistance, induced, at least in part, by the ATR-mediated DNA checkpoint pathway. Nevertheless, compelling evidence on the clinical efficacy of MTA1 inhibitors are still lacking and further studies on the clinical implementation of the drug are warranted. The development of relevant pharmacological inhibitors is also of importance.
Acknowledgments
The authors thank D. Yun Chen from Nanjing Medical University for providing the pLL-3.7 plasmids.
Funding: This work was supported by Wu Jieping Medical Foundation of the People’s Republic of China (No. 320.6750.18537), the National Natural Science Foundation of China (81772645), the Science and Technology Plan Project of Suzhou (SLT201913), Beijing Xisike Clinical Oncology Research Fundation (Y-XD2019-227), the Research Grant of BeiGene (COMM-86).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/atm-21-941
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-941
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-941). All authors report that this work was supported by Wu Jieping Medical Foundation of the People’s Republic of China (No. 320.6750.18537), the National Natural Science Foundation of China (81772645), the Science and Technology Plan Project of Suzhou (SLT201913), Beijing Xisike Clinical Oncology Research Fundation (Y-XD2019-227), the Research Grant of BeiGene (COMM-86). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Ethical Committee of the First Affiliated Hospital of Soochow University (Suzhou, China), and written informed consent was obtained from each patient. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- van Meerbeeck JP, Scherpereel A, Surmont VF, et al. Malignant pleural mesothelioma: the standard of care and challenges for future management. Crit Rev Oncol Hematol 2011;78:92-111. [Crossref] [PubMed]
- Bueno R, Opitz I. Surgery in Malignant Pleural Mesothelioma. J Thorac Oncol 2018;13:1638-54. [Crossref] [PubMed]
- Buikhuisen WA, Hiddinga BI, Baas P, et al. Second line therapy in malignant pleural mesothelioma: A systematic review. Lung Cancer 2015;89:223-31. [Crossref] [PubMed]
- Galluzzi L, Senovilla L, Vitale I, et al. Molecular mechanisms of cisplatin resistance. Oncogene 2012;31:1869-83. [Crossref] [PubMed]
- Köberle B, Tomicic MT, Usanova S, et al. Cisplatin resistance: preclinical findings and clinical implications. Biochim Biophys Acta 2010;1806:172-82. [PubMed]
- Furuta T, Ueda T, Aune G, et al. Transcription-coupled nucleotide excision repair as a determinant of cisplatin sensitivity of human cells. Cancer Res 2002;62:4899-902. [PubMed]
- Li Z, Pearlman AH, Hsieh P. DNA mismatch repair and the DNA damage response. DNA Repair (Amst) 2016;38:94-101. [Crossref] [PubMed]
- Vitale I, Galluzzi L, Castedo M, et al. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat Rev Mol Cell Biol 2011;12:385-92. [Crossref] [PubMed]
- Hurley PJ, Wilsker D, Bunz F. Human cancer cells require ATR for cell cycle progression following exposure to ionizing radiation. Oncogene 2007;26:2535-42. [Crossref] [PubMed]
- Jazayeri A, Falck J, Lukas C, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 2006;8:37-45. [Crossref] [PubMed]
- Pilié PG, Tang C, Mills GB, et al. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat Rev Clin Oncol 2019;16:81-104. [Crossref] [PubMed]
- Karnitz LM, Zou L. Molecular Pathways: Targeting ATR in Cancer Therapy. Clin Cancer Res 2015;21:4780-5. [Crossref] [PubMed]
- Lam MH, Rosen JM. Chk1 versus Cdc25: chking one's levels of cellular proliferation. Cell Cycle 2004;3:1355-7. [Crossref] [PubMed]
- Sørensen CS, Hansen LT, Dziegielewski J, et al. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol 2005;7:195-201. [Crossref] [PubMed]
- Wang X, Kennedy RD, Ray K, et al. Chk1-mediated phosphorylation of FANCE is required for the Fanconi anemia/BRCA pathway. Mol Cell Biol 2007;27:3098-108. [Crossref] [PubMed]
- Zhi G, Wilson JB, Chen X, et al. Fanconi anemia complementation group FANCD2 protein serine 331 phosphorylation is important for fanconi anemia pathway function and BRCA2 interaction. Cancer Res 2009;69:8775-83. [Crossref] [PubMed]
- Rundle S, Bradbury A, Drew Y, et al. Targeting the ATR-CHK1 Axis in Cancer Therapy. Cancers (Basel) 2017;9:41. [Crossref] [PubMed]
- Toh Y, Nicolson GL. The role of the MTA family and their encoded proteins in human cancers: molecular functions and clinical implications. Clin Exp Metastasis 2009;26:215-27. [Crossref] [PubMed]
- Kumar R, Wang RA. Structure, expression and functions of MTA genes. Gene 2016;582:112-21. [Crossref] [PubMed]
- Xu C, Hua F, Chen Y, et al. MTA1 promotes metastasis of MPM via suppression of E-cadherin. J Exp Clin Cancer Res 2015;34:151. [Crossref] [PubMed]
- Li DQ, Ohshiro K, Khan MN, et al. Requirement of MTA1 in ATR-mediated DNA damage checkpoint function. J Biol Chem 2010;285:19802-12. [Crossref] [PubMed]
- Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 2009;55:611-22. [Crossref] [PubMed]
- Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, et al. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 2004;73:39-85. [Crossref] [PubMed]
- Hans F, Dimitrov S. Histone H3 phosphorylation and cell division. Oncogene 2001;20:3021-7. [Crossref] [PubMed]
- Kindler HL, Ismaila N, Armato SG, et al. Treatment of Malignant Pleural Mesothelioma: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol 2018;36:1343-73. [Crossref] [PubMed]
- Mazurek JM, Syamlal G, Wood JM, et al. Malignant Mesothelioma Mortality - United States, 1999-2015. MMWR Morb Mortal Wkly Rep 2017;66:214-8. [Crossref] [PubMed]
- Abdel-Rahman O. Global trends in mortality from malignant mesothelioma: Analysis of WHO mortality database (1994-2013). Clin Respir J 2018;12:2090-100. [Crossref] [PubMed]
- Zalcman G, Mazieres J, Margery J, et al. Bevacizumab for newly diagnosed pleural mesothelioma in the Mesothelioma Avastin Cisplatin Pemetrexed Study (MAPS): a randomised, controlled, open-label, phase 3 trial. Lancet 2016;387:1405-14. [Crossref] [PubMed]
- Petrelli F, Ardito R, Conti B, et al. A systematic review and meta-analysis of second-line therapies for treatment of mesothelioma. Respir Med 2018;141:72-80. [Crossref] [PubMed]
- Guddeti RK, Bali P, Karyala P, et al. MTA1 coregulator regulates LDHA expression and function in breast cancer. Biochem Biophys Res Commun 2019;520:54-9. [Crossref] [PubMed]
- Toh Y, Ohga T, Endo K, et al. Expression of the metastasis-associated MTA1 protein and its relationship to deacetylation of the histone H4 in esophageal squamous cell carcinomas. Int J Cancer 2004;110:362-7. [Crossref] [PubMed]
- Kidd M, Modlin IM, Mane SM, et al. The role of genetic markers--NAP1L1, MAGE-D2, and MTA1--in defining small-intestinal carcinoid neoplasia. Ann Surg Oncol 2006;13:253-62. [Crossref] [PubMed]
- Kidd M, Modlin IM, Pfragner R, et al. Small bowel carcinoid (enterochromaffin cell) neoplasia exhibits transforming growth factor-beta1-mediated regulatory abnormalities including up-regulation of C-Myc and MTA1. Cancer 2007;109:2420-31. [Crossref] [PubMed]
- Malisetty VL, Penugurti V, Panta P, et al. MTA1 expression in human cancers - Clinical and pharmacological significance. Biomed Pharmacother 2017;95:956-64. [Crossref] [PubMed]
- Yi S, Guangqi H, Guoli H. The association of the expression of MTA1, nm23H1 with the invasion, metastasis of ovarian carcinoma. Chin Med Sci J 2003;18:87-92. [PubMed]
- Jia QP, Yan CY, Zheng XR, et al. Upregulation of MTA1 expression by human papillomavirus infection promotes CDDP resistance in cervical cancer cells via modulation of NF-κB/APOBEC3B cascade. Cancer Chemother Pharmacol 2019;83:625-37. [Crossref] [PubMed]
- Feng X, Zhang Q, Xia S, et al. MTA1 overexpression induces cisplatin resistance in nasopharyngeal carcinoma by promoting cancer stem cells properties. Mol Cells 2014;37:699-704. [Crossref] [PubMed]
- Qiu Z, Oleinick NL, Zhang J. ATR/CHK1 inhibitors and cancer therapy. Radiother Oncol 2018;126:450-64. [Crossref] [PubMed]
- Smith J, Tho LM, Xu N, et al. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res 2010;108:73-112. [Crossref] [PubMed]
- Prevo R, Fokas E, Reaper PM, et al. The novel ATR inhibitor VE-821 increases sensitivity of pancreatic cancer cells to radiation and chemotherapy. Cancer Biol Ther 2012;13:1072-81. [Crossref] [PubMed]
- Fokas E, Prevo R, Pollard JR, et al. Targeting ATR in vivo using the novel inhibitor VE-822 results in selective sensitization of pancreatic tumors to radiation. Cell Death Dis 2012;3:e441 [Crossref] [PubMed]
- Matthews DJ, Yakes FM, Chen J, et al. Pharmacological abrogation of S-phase checkpoint enhances the anti-tumor activity of gemcitabine in vivo. Cell Cycle 2007;6:104-10. [Crossref] [PubMed]
(English Language Editor: R. Scott)


