
A review of the application of machine learning in molecular imaging
Section 1: Introduction
The term of molecular imaging (MI) first came into use in the late 1990s, which applied various imaging techniques to display the changes of specific molecules in vivo and to conduct qualitative and quantitative studies on their biological behaviors at the level of tissue, cell and sub-cell (1). Compared with traditional medical imaging equipment, MI can effectively use specific molecular probes to monitor the process of tumor development at the molecular level in real time, laying a technical foundation for early detection, accurate diagnosis and effective treatment of diseases (2-4).
MI integrates molecular biochemistry, data processing, nanotechnology, image processing and other technologies to pursue high imaging specificity, sensitivity and resolution (5). Thus, MI is not a single or isolated technological innovation, but an integration and evolution of multidisciplinary technologies. MI modalities can be roughly categorized into three groups: classical anatomical imaging modalities [such as magnetic resonance imaging (MRI), X-ray computed tomography (CT), ultrasound imaging (USI), etc.], optical molecular imaging (OMI) modalities (such as bioluminescence imaging, fluorescence imaging, photoacoustic imaging, etc.), and nuclear medical imaging modalities [such as positron emission tomography (PET) and single photon emission computed tomography (SPECT)] (6,7). The classical anatomical imaging modalities have the longest history in equipment development, image processing and analysis, as well as biological and medical applications. A large number of literatures have reviewed the progress of these modalities in the field of MI (8-11). Therefore, the content of this paper mainly focuses on OMI and nuclear medical imaging technologies.
OMI becomes a new hot spot in medical imaging studies in recent years and has been widely applied in preclinical and clinical research. It uses molecular probes to label the target organism. Under certain external conditions, molecular probes emit fluorescence in the visible or near infrared spectrum, which can be detected by highly sensitive optical cameras. Then, the position and intensity of the fluorescent light source can be acquired to visualize the physiological activity information at the molecular and/or cellular level of the organism. Owing to the high sensitivity, no radiation, low cost, dynamic observation, and intuitive imaging, OMI has been widely used in tumor detection, drug development, surgical navigation, and many other fields (12-15). Furthermore, in order to obtain the three-dimensional (3D) information of the observed object, a variety of modern tomography techniques have been developed, such as the optical scattering tomography (OST) and photoacoustic tomography (PAT) (16-19). The corresponding imaging methods and hardware systems of OMI have also developed significantly.
Imaging diagnosis is an important part of clinical medicine, and nuclear medicine is playing an increasingly important role in imaging diagnosis. Nuclear medicine relies on radionuclides for imaging, including PET and SPECT. PET relies on positron-emitting nuclides for imaging, mainly including 18F, 68Ga, etc., while SPECT relies on single-photon-emitting nuclides and 99mTc is mainly used in clinical applications. Since PET and SPECT are important functional imaging methods with high sensitivity and specificity, they have been applied to tumors, central nervous system (CNS) and cardiovascular system (20-25). With the continuous development and progress of MI, PET and SPECT as important parts of MI have also made considerable progress in recent years.
Because of the rapid development of medical imaging technologies and the continuous expansion of medical data, the demand of accurate, automated and quantitative approaches for image processing and analysis becomes more and more urgent. Eventually, artificial intelligence (AI) has been widely applied in the field of MI. AI is a computer science that depends on the biological or physical coding. Furthermore, through human interventions, AI endows computer with a new respond in a similar way to human intelligence (26). Machine learning (ML) technology based on deep neural network (DNN), also known as deep learning (DL) method, is an important embodiment of AI (26). In recent years, benefiting from the rapid development of computer technology, the DL method is widely used to process large-scale and high-dimensional data. Owing to the powerful feature extraction and data processing performance, AI has been widely used in pattern recognition (27-31), natural language processing (32,33), network information security (34,35) and many other fields.
At present, the major applications of ML in MI can be divided into three parts: ML based imaging reconstruction (36-38), ML aided disease diagnosis (39,40), and intelligent target delineation (41,42). In the remainder of this article, we will introduce the relevant application of ML algorithms. Section 2 and 3 present the application of ML in OMI and nuclear medical imaging, respectively. In the section 4, we discuss some critical challenges and perspectives.
Section 2: Application of ML in OMI
In this section, we mainly introduce the recent applications of ML in fluorescence image-guided surgery (FIGS), OST, and PAT, which includes preclinical studies and clinical practices.
FIGS
As a highly sensitive and real-time intraoperative imaging technique, FIGS can assist operators to accurately and effectively locate and remove the malignant lesions in various kinds of clinical practices (Figure 1) (43-46). FIGS relies on fluorescence light produced by near-infrared (NIR) fluorescent contrast agents, which is strongly scattered and adsorbed by soft tissues, leading to limited but useful imaging depth. Because the optical scattering and absorption significantly affect imaging quality in FIGS, how to achieve better quality imaging of complex tissues in real-time remains a huge challenge (46-49). In recent years, many groups have been working on this issue. In terms of hardware, various imaging systems have been developed for intraoperative FIGS, including systems designed for open and endoscopic surgeries, as well as systems utilizing NIR-II signals (optical spectrum: 1,000–1,700 nm) (46,50-53). Besides those effects, the improvement of software with new imaging algorithms can further mitigate the physical limitations in hardware systems. One of the representative methods is the ML-based strategy, which has proven to be capable of enhancing the overall imaging quality frame-by-frame in intraoperative FIGS.
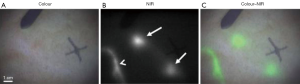
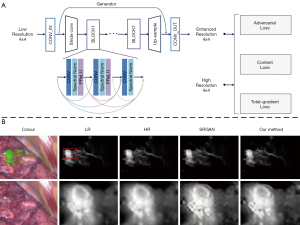
To improve imaging quality, ML is mainly used in image enhancement and image registration. Zhang et al. (47) proposed an image post-processing method by employing the generative adversarial network (GAN) to achieve image enhancement as shown in Figure 2A. In their study, a total gradient loss was presented for network training and a finetuning training procedure was applied into the network architecture. Therefore, the problem of fake texture caused by traditional neural networks was overcome and further enhancing the image resolution (Figure 2B). Ravì et al. (54) developed a synthetic data generation method conquer the lack of ground-truth data. This synthetic data was applied to train exemplar-based DNNs and obtained convincing super-resolution fluorescence image. Unger et al. (55) proposed a hybrid histological registration method to achieve accurate registration of autofluorescence imaging data with ex vivo histological images, which is meaning for evaluation of tumor margin.
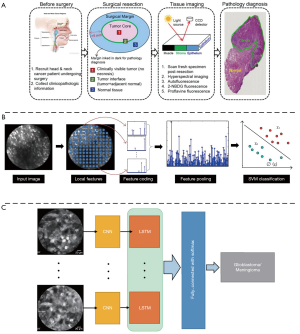
It is significant to determine the type and boundary of lesions accurately and quickly during the surgery, however, this is limited by the subjective consciousness of the operator and the delay of conventional surgical pathology. Therefore, in recent years, ML technology has been widely applied to intraoperative lesion analysis, which is expected to overcome the judgment errors caused by individuals and speed up the surgical pathology process by relying on automated technical analysis. Various groups performed quantitative analysis of fluorescence imaging of surgical specimens to determine tumor boundaries based on some traditional ML classifiers (SVM, random forest and so on) (56-58) (Figure 3A). These studies have achieved high accuracy, specificity and sensitivity (>90%), therefore holding potential for clinical surgery. In addition, there are also many researchers working on in vivo lesion analysis during the surgery. Using confocal laser endomicroscopy (CLE) imaging, Kamen et al. (59) proposed a ML-based algorithm to distinguish two types of brain tumors: glioblastoma and meningioma. They extracted the features of CLE images and using encoding schemes. Then SVM was used as a classifier and achieved an accuracy of more than 83% (Figure 3B). Li et al. (60) then proposed a video classification framework based on DL to classify glioblastoma and meningioma (Figure 3C). Under this framework, CNN was used to extract the features of each frame in the probe-based confocal laser endomicroscopy (pCLE) video, and then the RNN was used to fuse all the features. The results demonstrated that the proposed method improved the classification performance and achieved accuracy equal to 99.49%.
OST
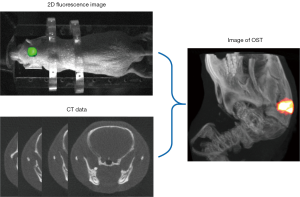
As a multimodal imaging technology, OST combines two-dimensional (2D) body surface fluorescence information with high-resolution structural imaging technology (MRI or CT). With the help of mathematical models, OST can accurately reconstruct the three-dimensional (3D) distribution of the internal light source from the fluorescence distribution on the surface (Figure 4), such as bioluminescence tomography (BLT) and fluorescence molecular tomography (FMT) (15,61,63). Compared with 2D fluorescence imaging, OST provides tomographic information and can quantify the 3D distribution of light source in the bodies, which is of great significance in preclinical research.
Most of conventional OST reconstruction algorithms are based on a photon propagation model of radiation transfer equation (RTE). Owing to the high complexity of RTE, a simplified low-order approximation model diffusion approximation equation (DE) is used to describe the photon propagation in biological tissues (64-66). Based on the finite element method, the distribution of fluorescence on the surface and internal light source is modeled as a linear relationship: AX = b. The reverse problem is to solve the light distribution X. However, OST suffers from the ill-posedness owing to the scattering effect, which influences the reconstruction accuracy (15,66,67). Therefore, various reconstruction strategies have been proposed to improve the reconstruction accuracy, such as regularization methods (L2, L1, LP) (17,68,69), Bayesian sparse based methods (62), matching pursuit algorithms (70,71), and guided methods utilizing the prior information of tumor segmentation (16,72). Nevertheless, the reconstruction performance of these model-based methods are still limited by ill-posedness and the model error caused by the simplified photon propagation model (37,73).
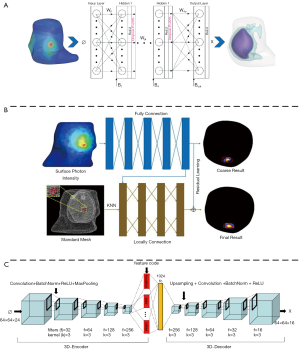
In recent years, many ML-based methods have been proposed to address the limitations of conventional model-based reconstruction algorithms. Gao et al. (37) developed a data-driven strategy based on the multi-layer perceptron (MLP) to achieve accurate location reconstruction in BLT as shown in Figure 5A. They utilized a Monte Carlo simulating method to collect training data. The error between the reconstructed light source and the real light source was used to train the network. As an extension, Meng et al. (73) connected a local connection sub-network based on the K-nearest neighbor (KNN-LC) over the fully connected network (FCN) and further improved the morphology reconstruction in FMT (Figure 5B). Further, owing to the large number of parameters required and the large amount of redundant information extracted for FCN, many researchers hope to apply convolutional neural network (CNN) to OST reconstruction. However, owing to the spatial topological structure of 3D mesh, CNN cannot be directly used for OST reconstruction. To address this problem, Li et al. (74) utilized graph convolution networks (GCN) in FMT reconstruction, which reduced the training parameters and achieved relatively rapid reconstruction. Moreover, Guo et al. (36) proposed a 3D deep encoder–decoder network to obtain the distribution of internal light source directly from 2D fluorescence image (Figure 5C). They collected 24 regular phantom surface 2D projection images from 0 to 360 degrees. Every image was resized to 64×64, thereby, the input format of the network was 64×64×24. And the output was 64×64×16. This strategy achieved FMT reconstruction directly from 2D fluorescence images that the limitations caused by finite element mesh could be avoided.
These ML-based methods address the model error caused by the photon propagation model and the ill-posedness caused by the inverse problem. However, there are still many limitations to be overcome in the practical application. One major limitation is that there is no unified public database for OST reconstruction. The training data are generated by simulations with individual differences, so it is lack of credibility. Furthermore, there is a lack of in vivo data. Thereby, a public database or a standard and effective data collection framework is urgently needed.
PAT
PAT is an emerging and noninvasive hybrid biomedical imaging technology that can be used to reveal the optical absorption characteristics of tissues (75-77). In PAT, pulsed laser is used to illuminate the biological tissues. Some of the absorbed light energy will be converted into thermal energy, causing nearby tissues to expand thermally, resulting in ultrasonic waves. The ultrasonic waves are measured by ultrasonic transducers and used for image reconstruction (78,79). The principle of PAT is shown in Figure 6. Since the optical absorption characteristics of biological tissues can reflect the hemoglobin concentration and molecular structure, PAT has great application potential in preclinical and clinical researches (80-82).

In the past decade, many researchers have made extensive efforts to improve the image reconstruction quality of PAT. In the field of conventional reconstruction algorithm, linear reconstruction algorithm is a simple and direct method which achieves the reconstruction from signal to image by solving a linear transformation. Furthermore, filtered back-projection (FBP) and time reversal method are two major types in linear reconstruction algorithms (83-87). Owing to their easy implementation and rapid reconstruction, they are widely applied in PAT reconstruction. However, the linear reconstruction algorithm will lead to many reconstructed artifacts in downsampled and limited-view data reconstruction (88,89). Another widely used conventional algorithm is model-based method, which relies on the photoacoustic forward propagation model (18,88,90-92). Model-based method is based on an iterative optimization framework with regularization prior. However, these methods are greatly affected by the noise caused by the photoacoustic signal acquisition, especially under the condition of downsampling, thereby the reconstruction results of these methods are limited (61,93).
In recent years, the ML-based methods have been proposed in PAT reconstruction to address the limitations of conventional reconstruction algorithms. The ML-based method can be divided into two parts, one of which relies on the conventional reconstruction algorithm and the other does not. For the first class, there are two main implementations: firstly, a U-Net network is used as a post-processing network to improve the coarse PAT images that are reconstructed by conventional reconstruction algorithms (79,94) (Figure 7A). Owing to the strong denoising ability of U-Net, this kind of algorithm is effective in eliminating image artifacts and denoising. Secondly, a neural network is utilized to simulate each iteration of conventional iterative algorithms. This strategy was first proposed by Hauptmann et al. (19). They designed a DNN to represent the iteration framework and introduced the gradient information of the photoacoustic data (Figure 7B). Boink et al. designed a CNN network based on partially learned algorithm to simultaneously achieve image reconstruction and segmentation of PAT (96). Compared with the strategy of using U-Net for post-processing network, the reconstruction results are further improved by the iterative network. These methods rely on conventional linear reconstruction algorithms to provide an initial value, and therefore, the reconstruction results are not stable enough. To address this limitation, it is necessary to achieve the direct reconstruction independent of conventional reconstruction methods. Waibel et al. first attempted to use CNN network to achieve this direct reconstruction, however, the results were not as good as the above strategies (97). Recently, Tong et al. proposed a DL network to improve the image quality directly from signal to image (95). They designed a Feature Projection Network (FPnet) to achieve domain transformation, and then a U-Net was used to further improve the image quality (Figure 7C). Their method is superior to some of the cutting-edge methods available.
Although the performance of the ML-based approach is superior to that of conventional algorithms, there are also limitations needed to be addressed. At present, most ML-based algorithms rely on conventional algorithms to provide an initial value, which makes this method not a real end-to-end DL algorithm. Besides, in the case of full view and dense sampling, the ML-based method does not have great advantages over the conventional method.
Section 3: Application of ML in nuclear medical imaging
In the past five years, the application of ML in clinical medicine, especially in nuclear medicine, has received increasing attention (98). With the rapid development in nuclear medicine and ML, the connection between ML and medical imaging is getting closer and closer (99,100). Image diagnosis is an important part of clinical work in nuclear medicine. During the diagnosis process, radiologist need to read a large number of image data and ML has made great progress in the reconstruction of CT, SPECT and PET (101,102). At the same time, ML is very helpful in assisting radiologists to diagnose and treat diseases (103-105).
Application of ML in PET
PET is an important part of nuclear medicine. The development of PET/CT has significantly improved the sensitivity and specificity of imaging diagnose and it is also of great significance in disease treatment and prognosis.
The combination of ML and imaging has been widely used in the detection and diagnosis of tumors (103). The diagnosis of lung nodules is an important part of the daily work of radiologists and how to diagnose the lung nodules is a tedious and complicated process. To date ML demonstrates remarkable and significant progress in pulmonary nodules detection and diagnosis (106-109). Nuclear medicine imaging needs to rely on radionuclides. 18F-FDG is the most commonly used radionuclide, but its imaging in the brain has limitations because of high background in normal brain. A variety of imaging agents based on the nervous system have been developed especially for CNS tumors. 18F-FET, a specific brain tumor imaging agent, significantly reduced the background of the normal brain, and improved the contrast of tumor. The combined application of CNN and 18F-FET PET/CT for the diagnosis of gliomas has obviously increased the sensitivity, specificity, positive predictive value and negative predictive value (110) (Figure 8). 11C-Choline PET/MRI has been shown to improve the diagnostic accuracy of primary prostate cancer. The application of PET/MR with ML algorithm has been tried to diagnose local prostate cancer (111). With the development of ML and new molecular probes, DNN has been used in 68Ga-PSMA imaging and made a better performance in the diagnosis of bone metastasis and lymph node metastasis from prostate cancer (112,113). Automated PET/CT segmentation trained with CNN was also used in the prostate cancer lesion uptake which was in association with overall survival (114,115). While PET/CT performs a whole body scan at one time, it can accurately detect systemic diseases, such as leukemia, lymphoma, multiple myeloma, etc. and then provide precision medicine (115-117). Nuclear alteration is a distinguished feature of many types of cancers and it is also an important evidence for pathological diagnosis. Recent studies based on ML approaches could automatically analyze data related to nuclear changes to assist diagnosis decision (118,119).
With the continuous research and development of molecular probes, PET combined with ML which plays an important role in the diagnosis of CNS tumor as mentioned above can also be applied to many other aspects of the nervous system. Alzheimer’s disease (AD) is the most prevalent form of age-related dementia that poses challenges to global health care systems (120). Many different types of molecular probes and different ML algorithms have been applied to the diagnosis of AD, cognitive impairment (MCI) and brain amyloid burden (121-128). Recently a ML model trained only by normal brain PET data was established and it could assist experts to identify and locate the abnormal patterns of PET images (129).
18F-FDG PET is also a common used mean of cardiac examination and it plays a fundamental role in diagnosis of cardiac sarcoidosis (CS) (25,130). A method based on deep convolutional neural network (DCNN) was proposed for CS classification (131).
The effective treatment and prognosis of the disease are the most concerned issues for patients, and ML can be used to guide the treatment plan, evaluate the tumor stage and predict the prognosis of the disease.
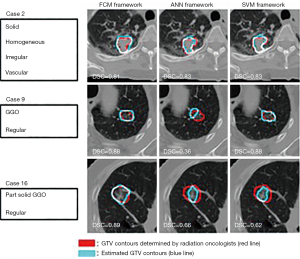
Radiotherapy and chemotherapy are important means of tumor treatment. PET/CT and other imaging modality can assist in the delineation of the target areas of radiotherapy and effectively monitor the tumor objective response rate (ORR) and metastasis (104,105,132,133). Stereotactic body radiation therapy (SBRT) which can precisely locate the tumor position and deliver a high dose to a tumor per fraction rather than to the surrounding normal tissue, is a promising technique for the treatment of cancer (134). Usually gross tumor volumes (GTVs) regions are manually delineated by radiation oncologist before the treatment and ML can be employed to help radiation oncologists to delineate GTV regions (135,136) (Figure 9). Lymph node metastasis and distant metastasis are the significant prognostic factor for cancer patients and the ability to predict them precisely is necessary for treatment optimization. With the assist of ML, PET/CT can accurately predict the staging and grading of tumors (137,138).
With the development of ML, some models have been established that can successfully predict and assess the response of chemoradiotherapy treatment in oncological patients (139-142). Although the diagnosis and treatment of the disease are critical to the patient, the survival rate of the tumor is the patient’s utmost concern. ML using in vivo, ex vivo, and patient features has made progress in predicting survival in 11C-MET-positive glioma patients (143) (Figure 10).
Recently, treatment strategies for AD are focusing on slowing cognitive decline. For the purpose of effective treatment of AD, it is important to identify subjects who are most likely to exhibit rapid cognitive decline (144,145). By applying CNN to FDG and AV-45 PET, a CNN-based method that could successfully predict cognitive decline was proposed (146). Several other in-depth learning algorithms combined with PET imaging were developed to predict cognitive performance and AD and it was very beneficial to the early treatment of patients (147-149).
Application of ML in SPECT
SPECT is one of the earliest nuclear medicine imaging equipment used in clinical applications. With the help of ML, SPECT has made great progress in the application of CNS and cardiovascular system.
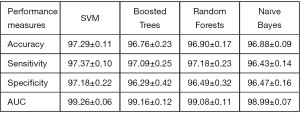
Parkinson disease (PD) is the most common neurological disorder disease with characteristic movement difficulty like tremor, stiffness and slowness. Dopamine transporter SPECT/CT has been proven to improve the diagnostic accuracy of Parkinson disease (150). The application of ML in SPECT makes PD’s diagnosis faster and helps physicians further improve the accuracy of diagnosis (151-153). For example, Prashanth et al. proposed that the support vector machine (SVM) classifier was applied to the recognition of PD. As shown in Figure 11, the accuracy could reach 97.29% (151).
Radionuclide myocardial perfusion imaging (MPI) which usually uses 99mTc-labeled tracers to map the relative distribution of myocardial blood flow (MBF) both at rest and with stress is the most commonly used detection method for heart disease (154,155). MPI has been used in the diagnosis of coronary artery disease (CAD) which is the leading cause of mortality worldwide and the artificial neural network (ANN) can help experts to detect myocardial perfusion defects and ischemia (156-159). Usually manual adjustment is required to accurately locate the position of mitral valve plane (VP) in the left ventricle of heart and Betancur et al. developed a ML method for fully automatic VP positioning in MPI without the need for expert intervention (160). One thousand patients underwent rest/stress SPECT MPI and the diagnostic performance of AI reporting system that generated a structured natural language report was comparable to the experts (161).
Heart-related diseases are one of the diseases with the highest fatality rate in China, and accurate prediction of related diseases and their risks is of great significance to treatment and prognosis. ML in combination with myocardial perfusion SPECT (MPS) has been developing rapidly and this approach is also applied to the prediction of cardiovascular diseases. Arsanjani et al. established a ML algorithm to predict early revascularization which was comparable to or better than the experienced experts (162). Trained with 1,638 patients (67% males) without known CAD, DL with MPI made better performance in the prediction of per-patient and per-vessel CAD compared with current clinical methods in 2018 (163). By integrating clinical and imaging data, ML could predict adverse cardiac events (MACE) risk in patients underwent SPECT MPI (164). With the aid of AI, the situation is changing that disease can be judged only by experts’ clinical experience.
Section 4: Challenges and perspectives
Although ML has achieved a significant breakthrough in clinical and preclinical applications, it still faces great challenges in theoretical research and clinical trials. Firstly, as we all know, ML-based algorithm as data-driven technology is greatly influenced by the amount of data. Compared with natural images, it is difficult for medical images to obtain enough data to build ML models. To overcome the limitation of clinical application data, building virtual or simulated data sets may be a practical and feasible solution in the short term. Future studies should focus on constantly improving the data volume, or developing ML technology based on small data sets. Secondly, although the application of ML can increase the reconstruction quality of OST, PAT, PET, etc., there are many concerns that ML will introduce more artifacts, which will affect the interpretation of images. What specific artifacts will be introduced into ML, and there is still a lack of research in this area. Thirdly, owing to the uninterpretability of neural networks, it is difficult to describe the imaging mechanisms of various imaging techniques, which is urgently needed to be overcome of neural networks. Further attempts should be made to explain the working principle of ML in different application scenarios through the visualization of model features and other methods. Lastly, the clinical diagnosis and other applications of ML assisted nuclear medical imaging are mostly retrospective data, lacking the validation of prospective multicentric clinical trials. This will need to be supplemented in the future.
Under the background of AI industrial revolution, imaging physicians should face the opportunity more objectively and positively. At present, AI technology is still in the initial stage, and it is more about a single image task to propose a solution, which is far away from the clinical work scene. The development of AI cannot be separated from doctors, and the work of doctors in the future cannot be separated from AI. Medical services assisted by machines will be the optimal solution in the path of diagnosis and treatment in the future.
Acknowledgments
Funding: The study was approved by the Ministry of Science and Technology of China under Grant No. 2017YFA0205200, 2017YFA0700401, and 2016YFC0103803; the National Natural Science Foundation of China under Grants No. 61671449, 81227901, and 81527805; and the Chinese Academy of Sciences under Grant No. GJJSTD20170004, KFJ-STS-ZDTP-059, YJKYYQ20180048, QYZDJ-SSW-JSC005, and XDBS01030200.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Dr. Steven P. Rowe) for the series “Artificial Intelligence in Molecular Imaging” published in Annals of Translational Medicine. The article has undergone external peer review.
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-5877
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5877). The series “Artificial Intelligence in Molecular Imaging” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Weissleder R. Molecular imaging: exploring the next frontier. Radiology 1999;212:609-14. [Crossref] [PubMed]
- Chiang A, Million RP. Personalized medicine in oncology: next generation. Nat Rev Drug Discov 2011;10:895-6. [Crossref] [PubMed]
- Scudellari M. Genomics contest underscores challenges of personalized medicine. Nat Med 2012;18:326. [Crossref] [PubMed]
- Zuckerman R, Milne CP. Market watch: industry perspectives on personalized medicine. Nat Rev Drug Discov 2012;11:178. [Crossref] [PubMed]
- Krenning EP, Valkema R, Kwekkeboom DJ, et al. Molecular imaging as in vivo molecular pathology for gastroenteropancreatic neuroendocrine tumors: Implications for follow-up after therapy. J Nucl Med 2005;46:76S-82S. [PubMed]
- Willmann JK, van Bruggen N, Dinkelborg LM, et al. Molecular imaging in drug development. Nat Rev Drug Discov 2008;7:591-607. [Crossref] [PubMed]
- James ML, Gambhir SS. A molecular imaging primer: modalities, imaging agents, and applications. Physiol Rev 2012;92:897-965. [Crossref] [PubMed]
- Higaki T, Nakamura Y, Tatsugami F, et al. Improvement of image quality at CT and MRI using deep learning. Jpn J Radiol 2019;37:73-80. [Crossref] [PubMed]
- Kolossváry M, De Cecco CN, Feuchtner G, et al. Advanced atherosclerosis imaging by CT: Radiomics, machine learning and deep learning. J Cardiovasc Comput Tomogr 2019;13:274-80. [Crossref] [PubMed]
- Biswas M, Kuppili V, Saba L, et al. State-of-the-art review on deep learning in medical imaging. Front Biosci (Landmark Ed) 2019;24:392-426. [Crossref] [PubMed]
- Brattain LJ, Telfer BA, Dhyani M, et al. Machine learning for medical ultrasound: status, methods, and future opportunities. Abdom Radiol (NY) 2018;43:786-99. [Crossref] [PubMed]
- Blankenberg FG, Strauss HW. Recent advances in the molecular imaging of programmed cell death: Part II—Non–probe-based MRI, ultrasound, and optical clinical imaging techniques. J Nucl Med 2013;54:1-4. [Crossref] [PubMed]
- Chi C, Du Y, Ye J, et al. Intraoperative imaging-guided cancer surgery: from current fluorescence molecular imaging methods to future multi-modality imaging technology. Theranostics 2014;4:1072-84. [Crossref] [PubMed]
- Hu Z, Qu Y, Wang K, et al. In vivo nanoparticle-mediated radiopharmaceutical-excited fluorescence molecular imaging. Nat Commun 2015;6:7560. [Crossref] [PubMed]
- Wang K, Chi CW, Hu ZH, et al. Optical Molecular Imaging Frontiers in Oncology: The Pursuit of Accuracy and Sensitivity. Engineering 2015;1:309-23. [Crossref]
- Davis SC, Samkoe KS, O'Hara JA, et al. Comparing implementations of magnetic-resonance-guided fluorescence molecular tomography for diagnostic classification of brain tumors. J Biomed Opt 2010;15:051602 [Crossref] [PubMed]
- Gao Y, Wang K, Jiang S, et al. Bioluminescence Tomography Based on Gaussian Weighted Laplace Prior Regularization forIn VivoMorphological Imaging of Glioma. IEEE Trans Med Imaging 2017;36:2343-54. [Crossref] [PubMed]
- Huang C, Wang K, Nie L, et al. Full-wave iterative image reconstruction in photoacoustic tomography with acoustically inhomogeneous media. IEEE Trans Med Imaging 2013;32:1097-110. [Crossref] [PubMed]
- Hauptmann A, Lucka F, Betcke M, et al. Model-Based Learning for Accelerated, Limited-View 3-D Photoacoustic Tomography. IEEE Trans Med Imaging 2018;37:1382-93. [Crossref] [PubMed]
- Ghafoor S, Burger IA, Vargas AH. Multimodality Imaging of Prostate Cancer. J Nucl Med 2019;60:1350-8. [Crossref] [PubMed]
- Graham MM, Metter DF. Evolution of nuclear medicine training: Past, present, and future. J Nucl Med 2007;48:257-68. [PubMed]
- Hope TA, Goodman JZ, Allen IE, et al. Metaanalysis of (68)Ga-PSMA-11 PET Accuracy for the Detection of Prostate Cancer Validated by Histopathology. J Nucl Med 2019;60:786-93. [Crossref] [PubMed]
- Meles SK, Teune LK, de Jong BM, et al. Metabolic Imaging in Parkinson Disease. J Nucl Med 2017;58:23-8. [Crossref] [PubMed]
- Czernin J, Sonni I, Razmaria A, et al. The Future of Nuclear Medicine as an Independent Specialty. J Nucl Med 2019;60:3S-12S. [Crossref] [PubMed]
- Ramirez R, Trivieri M, Fayad ZA, et al. Advanced Imaging in Cardiac Sarcoidosis. J Nucl Med 2019;60:892-8. [Crossref] [PubMed]
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436-44. [Crossref] [PubMed]
- Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Advances in neural information processing systems 2012;
- Farabet C, Couprie C, Najman L, et al. Learning hierarchical features for scene labeling. IEEE Trans Pattern Anal Mach Intell 2013;35:1915-29. [Crossref] [PubMed]
- Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions. Proceedings of the IEEE conference on computer vision and pattern recognition; 2015.
- Hinton G, Deng L, Yu D, et al. Deep neural networks for acoustic modeling in speech recognition: The shared views of four research groups. IEEE Signal Process Mag 2012;29:82-97. [Crossref]
- Sainath TN, Kingsbury B, Saon G, et al. Deep Convolutional Neural Networks for large-scale speech tasks. Neural Netw 2015;64:39-48. [Crossref] [PubMed]
- Bengio Y, Ducharme R, Vincent P, et al. A neural probabilistic language model. J Mach Learn Res 2003;3:1137-55.
- Sutskever I, Vinyals O, Le QV. Sequence to sequence learning with neural networks. Advances in neural information processing systems 2014;
- Quan WZ, Wang K, Yan DM, et al. Distinguishing Between Natural and Computer-Generated Images Using Convolutional Neural Networks. Ieee Transactions on Information Forensics and Security 2018;13:2772-87. [Crossref]
- Bayar B, Stamm MC. Constrained Convolutional Neural Networks: A New Approach Towards General Purpose Image Manipulation Detection. Ieee Transactions on Information Forensics and Security 2018;13:2691-706. [Crossref]
- Guo L, Liu F, Cai C, et al. 3D deep encoder–decoder network for fluorescence molecular tomography. Opt Lett 2019;44:1892-5. [Crossref] [PubMed]
- Gao Y, Wang K, An Y, et al. Nonmodel-based bioluminescence tomography using a machine-learning reconstruction strategy. Optica 2018;5:1451-4. [Crossref]
- Davoudi N, Deán-Ben XL, Razansky D. Deep learning optoacoustic tomography with sparse data. Nat Mach Intell 2019;1:453-60. [Crossref]
- Ahmed MR, Zhang Y, Feng Z, et al. Neuroimaging and machine learning for dementia diagnosis: Recent advancements and future prospects. IEEE Rev Biomed Eng 2019;12:19-33. [Crossref] [PubMed]
- Nensa F, Demircioglu A, Rischpler C. Artificial Intelligence in Nuclear Medicine. J Nucl Med 2019;60:29S-37S. [Crossref] [PubMed]
- Giraud P, Giraud P, Gasnier A, et al. Radiomics and Machine Learning for Radiotherapy in Head and Neck Cancers. Front Oncol 2019;9:174. [Crossref] [PubMed]
- Sahiner B, Pezeshk A, Hadjiiski LM, et al. Deep learning in medical imaging and radiation therapy. Med Phys 2019;46:e1-e36. [Crossref] [PubMed]
- van der Vorst J, Schaafsma B, Verbeek F, et al. Dose optimization for near‐infrared fluorescence sentinel lymph node mapping in patients with melanoma. Br J Dermatol 2013;168:93-8. [Crossref] [PubMed]
- Gioux S, Choi HS, Frangioni JV. Image-Guided Surgery Using Invisible Near-Infrared Light: Fundamentals of Clinical Translation. Molecular Imaging 2010;9:237-55. [Crossref] [PubMed]
- Schaafsma BE, Mieog JSD, Hutteman M, et al. The clinical use of indocyanine green as a near-infrared fluorescent contrast agent for image-guided oncologic surgery. J Surg Oncol 2011;104:323-32. [Crossref] [PubMed]
- Hu Z, Fang C, Li B, et al. First-in-human liver-tumour surgery guided by multispectral fluorescence imaging in the visible and near-infrared-I/II windows. Nat Biomed Eng 2020;4:259-71. [Crossref] [PubMed]
- Zhang C, Wang K, An Y, et al. Improved generative adversarial networks using the total gradient loss for the resolution enhancement of fluorescence images. Biomed Opt Express 2019;10:4742-56. [Crossref] [PubMed]
- Zdankowski P, McGloin D, Swedlow JR. Full volume super-resolution imaging of thick mitotic spindle using 3D AO STED microscope. Biomedical Optics Express 2019;10:1999-2009. [Crossref] [PubMed]
- Vahrmeijer AL, Hutteman M, van der Vorst JR, et al. Image-guided cancer surgery using near-infrared fluorescence. Nat Rev Clin Oncol 2013;10:507-18. [Crossref] [PubMed]
- Garcia M, Edmiston C, York T, et al. Bio-inspired imager improves sensitivity in near-infrared fluorescence image-guided surgery. Optica 2018;5:413-22. [Crossref] [PubMed]
- Takada M, Takeuchi M, Suzuki E, et al. Real-time navigation system for sentinel lymph node biopsy in breast cancer patients using projection mapping with indocyanine green fluorescence. Breast Cancer 2018;25:650-5. [Crossref] [PubMed]
- Venugopal V, Park M, Ashitate Y, et al. Design and characterization of an optimized simultaneous color and near-infrared fluorescence rigid endoscopic imaging system. J Biomed Opt 2013;18:126018 [Crossref] [PubMed]
- Volpi D, Tullis IDC, Barber PR, et al. Electrically tunable fluidic lens imaging system for laparoscopic fluorescence-guided surgery. Biomed Opt Express 2017;8:3232-47. [Crossref] [PubMed]
- Ravì D, Szczotka AB, Shakir DI, et al. Effective deep learning training for single-image super-resolution in endomicroscopy exploiting video-registration-based reconstruction. Int J Comput Assist Radiol Surg 2018;13:917-24. [Crossref] [PubMed]
- Unger J, Sun T, Chen YL, et al. Method for accurate registration of tissue autofluorescence imaging data with corresponding histology: a means for enhanced tumor margin assessment. J Biomed Opt 2018;23:1-11. [Crossref] [PubMed]
- Unger J, Hebisch C, Phipps JE, et al. Real-time diagnosis and visualization of tumor margins in excised breast specimens using fluorescence lifetime imaging and machine learning. Biomed Opt Express 2020;11:1216-30. [Crossref] [PubMed]
- Lu G, Little JV, Wang X, et al. Detection of Head and Neck Cancer in Surgical Specimens Using Quantitative Hyperspectral Imaging. Clin Cancer Res 2017;23:5426-36. [Crossref] [PubMed]
- Fei B, Lu G, Wang X, et al. Label-free reflectance hyperspectral imaging for tumor margin assessment: a pilot study on surgical specimens of cancer patients. J Biomed Opt 2017;22:1-7. [Crossref] [PubMed]
- Kamen A, Sun S, Wan S, et al. Automatic Tissue Differentiation Based on Confocal Endomicroscopic Images for Intraoperative Guidance in Neurosurgery. Biomed Res Int 2016;2016:6183218 [Crossref] [PubMed]
- Li Y, Charalampaki P, Liu Y, et al. Context aware decision support in neurosurgical oncology based on an efficient classification of endomicroscopic data. Int J Comput Assist Radiol Surg 2018;13:1187-99. [Crossref] [PubMed]
- An Y, Meng H, Gao Y, et al. Application of machine learning method in optical molecular imaging: a review. Science China-Information Sciences 2020;63:111101 [Crossref]
- Yin L, Wang K, Tong T, et al. Improved Block Sparse Bayesian Learning Method Using K-Nearest Neighbor Strategy for Accurate Tumor Morphology Reconstruction in Bioluminescence Tomography. IEEE Trans Biomed Eng 2020;67:2023-32. [PubMed]
- Ntziachristos V, Ripoll J, Wang LV, et al. Looking and listening to light: the evolution of whole-body photonic imaging. Nat Biotechnol 2005;23:313-20. [Crossref] [PubMed]
- Arridge SR, Schweiger M, Hiraoka M, et al. A finite element approach for modeling photon transport in tissue. Med Phys 1993;20:299-309. [Crossref] [PubMed]
- Arridge SR. Optical tomography in medical imaging. Inverse Probl 1999;15:R41-R93. [Crossref]
- Wang G, Li Y, Jiang M. Uniqueness theorems in bioluminescence tomography. Med Phys 2004;31:2289-99. [Crossref] [PubMed]
- Qin C, Zhu SP, Feng JC, et al. Comparison of permissible source region and multispectral data using efficient bioluminescence tomography method. J Biophotonics 2011;4:824-39. [Crossref] [PubMed]
- Lu Y, Zhang X, Douraghy A, et al. Source reconstruction for spectrally-resolved bioluminescence tomography with sparse a priori information. Opt Express 2009;17:8062-80. [Crossref] [PubMed]
- Liu K, Tian J, Qin C, et al. Tomographic bioluminescence imaging reconstruction via a dynamically sparse regularized global method in mouse models. J Biomed Opt 2011;16:046016 [Crossref] [PubMed]
- Chehade M, Srivastava AK, Bulte JW. Co-Registration of Bioluminescence Tomography, Computed Tomography, and Magnetic Resonance Imaging for Multimodal In Vivo Stem Cell Tracking. Tomography 2016;2:159-65. [Crossref] [PubMed]
- Zhang X, Lu Y, Chan T. A novel sparsity reconstruction method from Poisson data for 3D bioluminescence tomography. J Sci Comput 2012;50:519-35. [Crossref]
- Davis SC, Samkoe KS, Tichauer KM, et al. Dynamic dual-tracer MRI-guided fluorescence tomography to quantify receptor density in vivo. Proc Natl Acad Sci U S A 2013;110:9025-30. [Crossref] [PubMed]
- Meng H, Gao Y, Yang X, et al. K-nearest Neighbor Based Locally Connected Network for Fast Morphological Reconstruction in Fluorescence Molecular Tomography. IEEE Trans Med Imaging 2020;39:3019-28. [Crossref] [PubMed]
- Li DS, Chen CX, Li JF, et al. Reconstruction of fluorescence molecular tomography based on graph convolution networks. Journal of Optics 2020;22:045602 [Crossref]
- Wang LV. Multiscale photoacoustic microscopy and computed tomography. Nat Photonics 2009;3:503-9. [Crossref] [PubMed]
- Kruger RA, Liu P, Fang YR, et al. Photoacoustic ultrasound (PAUS)—reconstruction tomography. Med Phys 1995;22:1605-9. [Crossref] [PubMed]
- Karabutov AA, Podymova NB, Letokhov VS. Time-resolved laser optoacoustic tomography of inhomogeneous media. Applied Physics B-Lasers and Optics 1996;63:545-63. [Crossref]
- Ntziachristos V, Razansky D. Molecular imaging by means of multispectral optoacoustic tomography (MSOT). Chem Rev 2010;110:2783-94. [Crossref] [PubMed]
- Antholzer S, Haltmeier M, Schwab J. Deep learning for photoacoustic tomography from sparse data. Inverse Probl Sci Eng 2018;27:987-1005. [Crossref] [PubMed]
- Zhang J, Chen B, Zhou M, et al. Photoacoustic image classification and segmentation of breast cancer: A feasibility study. IEEE Access 2018;7:5457-66.
- Wang LV, Hu S. Photoacoustic tomography: in vivo imaging from organelles to organs. Science 2012;335:1458-62. [Crossref] [PubMed]
- Rajanna AR, Ptucha R, Sinha S, et al. Prostate cancer detection using photoacoustic imaging and deep learning. Electronic Imaging 2016;2016:1-6. [Crossref]
- Xu M, Wang LV. Universal back-projection algorithm for photoacoustic computed tomography. Phys Rev E Stat Nonlin Soft Matter Phys 2005;71:016706 [Crossref] [PubMed]
- Burgholzer P, Bauer-Marschallinger J, Grun H, et al. Temporal back-projection algorithms for photoacoustic tomography with integrating line detectors. Inverse Problems 2007;23:S65-S80. [Crossref]
- Zeng L, Da X, Gu H, et al. High antinoise photoacoustic tomography based on a modified filtered backprojection algorithm with combination wavelet. Med Phys 2007;34:556-63. [Crossref] [PubMed]
- Xu Y, Wang LV. Time reversal and its application to tomography with diffracting sources. Phys Rev Lett 2004;92:033902 [Crossref] [PubMed]
- Hristova Y, Kuchment P, Nguyen L. Reconstruction and time reversal in thermoacoustic tomography in acoustically homogeneous and inhomogeneous media. Inverse Probl 2008;24:055006 [Crossref]
- Rosenthal A, Razansky D, Ntziachristos V. Fast semi-analytical model-based acoustic inversion for quantitative optoacoustic tomography. IEEE Trans Med Imaging 2010;29:1275-85. [Crossref] [PubMed]
- Paltauf G, Viator JA, Prahl SA, et al. Iterative reconstruction algorithm for optoacoustic imaging. J Acoust Soc Am 2002;112:1536-44. [Crossref] [PubMed]
- Dean-Ben XL, Ntziachristos V, Razansky D. Acceleration of Optoacoustic Model-Based Reconstruction Using Angular Image Discretization. IEEE Trans Med Imaging 2012;31:1154-62. [Crossref] [PubMed]
- Deán-Ben XL, Buehler A, Ntziachristos V, et al. Accurate model-based reconstruction algorithm for three-dimensional optoacoustic tomography. IEEE Trans Med Imaging 2012;31:1922-8. [Crossref] [PubMed]
- Arridge SR, Betcke MM, Cox BT, et al. On the adjoint operator in photoacoustic tomography. Inverse Probl 2016;32:115012 [Crossref]
- Arridge S, Beard P, Betcke M, et al. Accelerated high-resolution photoacoustic tomography via compressed sensing. Phys Med Biol 2016;61:8908-40. [Crossref] [PubMed]
- Schwab J, Antholzer S, Nuster R, et al. Real-time photoacoustic projection imaging using deep learning. arXiv preprint arXiv:06693 2018.
- Tong T, Huang W, Wang K, et al. Domain Transform Network for Photoacoustic Tomography from Limited-view and Sparsely Sampled Data. Photoacoustics 2020;19:100190 [Crossref] [PubMed]
- Boink YE, Manohar S, Brune C. A Partially-Learned Algorithm for Joint Photo-acoustic Reconstruction and Segmentation. IEEE Trans Med Imaging 2020;39:129-39. [Crossref] [PubMed]
- Waibel D, Gröhl J, Isensee F, et al., editors. Reconstruction of initial pressure from limited view photoacoustic images using deep learning. Photons Plus Ultrasound: Imaging and Sensing 2018; 2018: International Society for Optics and Photonics.
- Larvie M. Machine Learning in Radiology: Resistance Is Futile. Radiology 2019;290:465-6. [Crossref] [PubMed]
- Uribe CF, Mathotaarachchi S, Gaudet V, et al. Machine Learning in Nuclear Medicine: Part 1-Introduction. J Nucl Med 2019;60:451-8. [Crossref] [PubMed]
- Mayerhoefer ME, Materka A, Langs G, et al. Introduction to Radiomics. J Nucl Med 2020;61:488-95. [Crossref] [PubMed]
- Wang Y, Ma G, An L, et al. Semisupervised Tripled Dictionary Learning for Standard-Dose PET Image Prediction Using Low-Dose PET and Multimodal MRI. IEEE Trans Biomed Eng 2017;64:569-79. [Crossref] [PubMed]
- Hainc N, Federau C, Stieltjes B, et al. The Bright, Artificial Intelligence-Augmented Future of Neuroimaging Reading. Front Neurol 2017;8:489. [Crossref] [PubMed]
- Hustinx R. Physician centred imaging interpretation is dying out - why should I be a nuclear medicine physician? Eur J Nucl Med Mol Imaging 2019;46:2708-14. [Crossref] [PubMed]
- Nestle U, Schimek-Jasch T, Kremp S, et al. Imaging-based target volume reduction in chemoradiotherapy for locally advanced non-small-cell lung cancer (PET-Plan): a multicentre, open-label, randomised, controlled trial. Lancet Oncol 2020;21:581-92. [Crossref] [PubMed]
- Noordman BJ, Spaander MCW, Valkema R, et al. Detection of residual disease after neoadjuvant chemoradiotherapy for oesophageal cancer (preSANO): a prospective multicentre, diagnostic cohort study. Lancet Oncol 2018;19:965-74. [Crossref] [PubMed]
- Kawagishi M, Chen B, Furukawa D, et al. A study of computer-aided diagnosis for pulmonary nodule: comparison between classification accuracies using calculated image features and imaging findings annotated by radiologists. Int J Comput Assist Radiol Surg 2017;12:767-76. [Crossref] [PubMed]
- Schwyzer M, Martini K, Benz DC, et al. Artificial intelligence for detecting small FDG-positive lung nodules in digital PET/CT: impact of image reconstructions on diagnostic performance. Eur Radiol 2020;30:2031-40. [Crossref] [PubMed]
- Liu B, Chi W, Li X, et al. Evolving the pulmonary nodules diagnosis from classical approaches to deep learning-aided decision support: three decades' development course and future prospect. J Cancer Res Clin Oncol 2020;146:153-85. [Crossref] [PubMed]
- Schwyzer M, Ferraro DA, Muehlematter UJ, et al. Automated detection of lung cancer at ultralow dose PET/CT by deep neural networks - Initial results. Lung Cancer 2018;126:170-3. [Crossref] [PubMed]
- Blanc-Durand P, Van Der Gucht A, Schaefer N, et al. Automatic lesion detection and segmentation of 18F-FET PET in gliomas: A full 3D U-Net convolutional neural network study. PLoS One 2018;13:e0195798 [Crossref] [PubMed]
- Gatidis S, Scharpf M, Martirosian P, et al. Combined unsupervised-supervised classification of multiparametric PET/MRI data: application to prostate cancer. NMR Biomed 2015;28:914-22. [Crossref] [PubMed]
- Zhao Y, Gafita A, Vollnberg B, et al. Deep neural network for automatic characterization of lesions on (68)Ga-PSMA-11 PET/CT. Eur J Nucl Med Mol Imaging 2020;47:603-13. [Crossref] [PubMed]
- Zhao Y, Gafita A, Tetteh G, et al. Deep Neural Network for Automatic Characterization of Lesions on 68Ga-PSMA PET/CT Images. Conference proceedings: Annual International Conference of the IEEE Engineering in Medicine and Biology Society IEEE Engineering in Medicine and Biology Society Annual Conference 2019;2019:951-4.
- Polymeri E, Sadik M, Kaboteh R, et al. Deep learning-based quantification of PET/CT prostate gland uptake: association with overall survival. Clin Physiol Funct Imaging 2020;40:106-13. [Crossref] [PubMed]
- Mortensen MA, Borrelli P, Poulsen MH, et al. Artificial intelligence-based versus manual assessment of prostate cancer in the prostate gland: a method comparison study. Clin Physiol Funct Imaging 2019;39:399-406. [Crossref] [PubMed]
- Lee SI, Celik S, Logsdon BA, et al. A machine learning approach to integrate big data for precision medicine in acute myeloid leukemia. Nat Commun 2018;9:42. [Crossref] [PubMed]
- Xu L, Tetteh G, Lipkova J, et al. Automated Whole-Body Bone Lesion Detection for Multiple Myeloma on (68)Ga-Pentixafor PET/CT Imaging Using Deep Learning Methods. Contrast Media Mol Imaging 2018;2018:2391925 [Crossref] [PubMed]
- Litjens G, Sanchez CI, Timofeeva N, et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep 2016;6:26286. [Crossref] [PubMed]
- Carleton NM, Lee G, Madabhushi A, et al. Advances in the computational and molecular understanding of the prostate cancer cell nucleus. J Cell Biochem 2018;119:7127-42. [Crossref] [PubMed]
- Katako A, Shelton P, Goertzen AL, et al. Machine learning identified an Alzheimer's disease-related FDG-PET pattern which is also expressed in Lewy body dementia and Parkinson's disease dementia. Sci Rep 2018;8:13236. [Crossref] [PubMed]
- Cattell L, Platsch G, Pfeiffer R, et al. Classification of amyloid status using machine learning with histograms of oriented 3D gradients. Neuroimage Clin 2016;12:990-1003. [Crossref] [PubMed]
- Suk HI, Lee SW, Shen D, et al. Deep sparse multi-task learning for feature selection in Alzheimer's disease diagnosis. Brain Struct Funct 2016;221:2569-87. [Crossref] [PubMed]
- Liu M, Zhang J, Yap PT, et al. Diagnosis of Alzheimer's Disease Using View-Aligned Hypergraph Learning with Incomplete Multi-modality Data. Med Image Comput Comput Assist Interv 2016;9900:308-16.
- Kang SK, Seo S, Shin SA, et al. Adaptive template generation for amyloid PET using a deep learning approach. Hum Brain Mapp 2018;39:3769-78. [Crossref] [PubMed]
- Lu D, Popuri K, Ding GW, et al. Multimodal and Multiscale Deep Neural Networks for the Early Diagnosis of Alzheimer's Disease using structural MR and FDG-PET images. Sci Rep 2018;8:5697. [Crossref] [PubMed]
- Zukotynski K, Gaudet V, Kuo PH, et al. The Use of Random Forests to Classify Amyloid Brain PET. Clin Nucl Med 2019;44:784-8. [Crossref] [PubMed]
- Li F, Tran L, Thung KH, et al. A Robust Deep Model for Improved Classification of AD/MCI Patients. IEEE J Biomed Health Inform 2015;19:1610-6. [Crossref] [PubMed]
- Kim J, Lee B. Identification of Alzheimer's disease and mild cognitive impairment using multimodal sparse hierarchical extreme learning machine. Hum Brain Mapp 2018;39:3728-41. [Crossref] [PubMed]
- Choi H, Ha S, Kang H, et al. Deep learning only by normal brain PET identify unheralded brain anomalies. EBioMedicine 2019;43:447-53. [Crossref] [PubMed]
- Youssef G, Leung E, Mylonas I, et al. The use of 18F-FDG PET in the diagnosis of cardiac sarcoidosis: a systematic review and metaanalysis including the Ontario experience. J Nucl Med 2012;53:241-8. [Crossref] [PubMed]
- Togo R, Hirata K, Manabe O, et al. Cardiac sarcoidosis classification with deep convolutional neural network-based features using polar maps. Comput Biol Med 2019;104:81-6. [Crossref] [PubMed]
- Mehanna H, Wong WL, McConkey CC, et al. PET-CT Surveillance versus Neck Dissection in Advanced Head and Neck Cancer. N Engl J Med 2016;374:1444-54. [Crossref] [PubMed]
- Aykan NF, Ozatli T. Objective response rate assessment in oncology: Current situation and future expectations. World J Clin Oncol 2020;11:53-73. [Crossref] [PubMed]
- Cao C, Huang J, Rimner A, et al. Stereotactic Body Radiation Therapy: Focusing on the Short Game. J Clin Oncol 2018;36:2455-6. [Crossref] [PubMed]
- Berthon B, Marshall C, Evans M, et al. ATLAAS: an automatic decision tree-based learning algorithm for advanced image segmentation in positron emission tomography. Phys Med Biol 2016;61:4855-69. [Crossref] [PubMed]
- Kawata Y, Arimura H, Ikushima K, et al. Impact of pixel-based machine-learning techniques on automated frameworks for delineation of gross tumor volume regions for stereotactic body radiation therapy. Phys Med 2017;42:141-9. [Crossref] [PubMed]
- Madekivi V, Bostrom P, Karlsson A, et al. Can a machine-learning model improve the prediction of nodal stage after a positive sentinel lymph node biopsy in breast cancer? Acta Oncol 2020;59:689-95. [Crossref] [PubMed]
- Zhou Z, Liyuan C, Sher D, et al. Predicting Lymph Node Metastasis in Head and Neck Cancer by Combining Many-objective Radiomics and 3-dimensioal Convolutional Neural Network through Evidential Reasoning. Annu Int Conf IEEE Eng Med Biol Soc 2018;2018:1-4. [Crossref] [PubMed]
- Ypsilantis PP, Siddique M, Sohn HM, et al. Predicting Response to Neoadjuvant Chemotherapy with PET Imaging Using Convolutional Neural Networks. PLoS One 2015;10:e0137036 [Crossref] [PubMed]
- Nogueira MA, Abreu PH, Martins P, et al. An artificial neural networks approach for assessment treatment response in oncological patients using PET/CT images. BMC Med Imaging 2017;17:13. [Crossref] [PubMed]
- Xiong J, Yu W, Ma J, et al. The Role of PET-Based Radiomic Features in Predicting Local Control of Esophageal Cancer Treated with Concurrent Chemoradiotherapy. Sci Rep 2018;8:9902. [Crossref] [PubMed]
- Buizza G, Toma-Dasu I, Lazzeroni M, et al. Early tumor response prediction for lung cancer patients using novel longitudinal pattern features from sequential PET/CT image scans. Phys Med 2018;54:21-9. [Crossref] [PubMed]
- Papp L, Pötsch N, Grahovac M, et al. Glioma Survival Prediction with Combined Analysis of In Vivo11C-MET PET Features, Ex Vivo Features, and Patient Features by Supervised Machine Learning. J Nucl Med 2018;59:892-9. [Crossref] [PubMed]
- Petersen RC. Mild cognitive impairment as a diagnostic entity. J Intern Med 2004;256:183-94. [Crossref] [PubMed]
- Petersen RC, Smith GE, Waring SC, et al. Mild cognitive impairment - Clinical characterization and outcome. Arch Neurol 1999;56:303-8. [Crossref] [PubMed]
- Choi H, Jin KHAlzheimer’s Disease Neuroimaging Initiative. Predicting cognitive decline with deep learning of brain metabolism and amyloid imaging. Behav Brain Res 2018;344:103-9. [Crossref] [PubMed]
- Ding Y, Sohn JH, Kawczynski MG, et al. A Deep Learning Model to Predict a Diagnosis of Alzheimer Disease by Using 18F-FDG PET of the Brain. Radiology 2019;290:456-64. [Crossref] [PubMed]
- Kim JP, Kim J, Kim Y, et al. Staging and quantification of florbetaben PET images using machine learning: impact of predicted regional cortical tracer uptake and amyloid stage on clinical outcomes. Eur J Nucl Med Mol Imaging 2020;47:1971-83. [Crossref] [PubMed]
- Schütze M, de Souza Costa D, de Paula JJ, et al. Use of machine learning to predict cognitive performance based on brain metabolism in Neurofibromatosis type 1. PLoS One 2018;13:e0203520 [Crossref] [PubMed]
- Armstrong MJ, Okun MS. Diagnosis and Treatment of Parkinson Disease: A Review. JAMA 2020;323:548-60. [Crossref] [PubMed]
- Prashanth R, Roy SD, Mandal PK, et al. High-Accuracy Classification of Parkinson's Disease Through Shape Analysis and Surface Fitting in 123I-Ioflupane SPECT Imaging. IEEE J Biomed Health Inform 2017;21:794-802. [Crossref] [PubMed]
- Choi H, Ha S, Im HJ, et al. Refining diagnosis of Parkinson's disease with deep learning-based interpretation of dopamine transporter imaging. Neuroimage Clin 2017;16:586-94. [Crossref] [PubMed]
- Kim DH, Wit H, Thurston M. Artificial intelligence in the diagnosis of Parkinson's disease from ioflupane-123 single-photon emission computed tomography dopamine transporter scans using transfer learning. Nucl Med Commun 2018;39:887-93. [Crossref] [PubMed]
- Gewirtz H, Dilsizian V. Integration of Quantitative Positron Emission Tomography Absolute Myocardial Blood Flow Measurements in the Clinical Management of Coronary Artery Disease. Circulation 2016;133:2180-96. [Crossref] [PubMed]
- Levsky JM, Spevack DM, Travin MI, et al. Coronary Computed Tomography Angiography Versus Radionuclide Myocardial Perfusion Imaging in Patients With Chest Pain Admitted to Telemetry: A Randomized Trial. Ann Intern Med 2015;163:174-83. [Crossref] [PubMed]
- Nakajima K, Matsuo S, Wakabayashi H, et al. Diagnostic Performance of Artificial Neural Network for Detecting Ischemia in Myocardial Perfusion Imaging. Circ J 2015;79:1549-56. [Crossref] [PubMed]
- Nakajima K, Kudo T, Nakata T, et al. Diagnostic accuracy of an artificial neural network compared with statistical quantitation of myocardial perfusion images: a Japanese multicenter study. Eur J Nucl Med Mol Imaging 2017;44:2280-9. [Crossref] [PubMed]
- Nakajima K, Okuda K, Watanabe S, et al. Artificial neural network retrained to detect myocardial ischemia using a Japanese multicenter database. Ann Nucl Med 2018;32:303-10. [Crossref] [PubMed]
- Shibutani T, Nakajima K, Wakabayashi H, et al. Accuracy of an artificial neural network for detecting a regional abnormality in myocardial perfusion SPECT. Ann Nucl Med 2019;33:86-92. [Crossref] [PubMed]
- Betancur J, Rubeaux M, Fuchs TA, et al. Automatic Valve Plane Localization in Myocardial Perfusion SPECT/CT by Machine Learning: Anatomic and Clinical Validation. J Nucl Med 2017;58:961-7. [Crossref] [PubMed]
- Garcia EV, Klein JL, Moncayo V, et al. Diagnostic performance of an artificial intelligence-driven cardiac-structured reporting system for myocardial perfusion SPECT imaging. J Nucl Cardiol 2020;27:1652-64. [Crossref] [PubMed]
- Arsanjani R, Dey D, Khachatryan T, et al. Prediction of revascularization after myocardial perfusion SPECT by machine learning in a large population. J Nucl Cardiol 2015;22:877-84. [Crossref] [PubMed]
- Betancur J, Commandeur F, Motlagh M, et al. Deep Learning for Prediction of Obstructive Disease From Fast Myocardial Perfusion SPECT. JACC Cardiovasc Imaging 2018;11:1654-63. [Crossref] [PubMed]
- Betancur J, Otaki Y, Motwani M, et al. Prognostic Value of Combined Clinical and Myocardial Perfusion Imaging Data Using Machine Learning. JACC Cardiovasc Imaging 2018;11:1000-9. [Crossref] [PubMed]



