
LncRNA prostate androgen-regulated transcript 1 (PART 1) functions as an oncogene in osteosarcoma via sponging miR-20b-5p to upregulate BAMBI
Introduction
Osteosarcoma (OS) is a bone malignancy originating from osteoid bone tissue which has a high mortality rate (1,2). Despite the effectiveness of therapeutic modalities for OS, including surgical resection, neoadjuvant, or adjuvant radiotherapy or chemotherapy, the rates of mortality and metastasis among patients with the disease are still extremely high. Moreover, for patients with metastatic or recurrent disease, the survival outlook is dismal (3,4). Multiple pathophysiological and pathological processes, such as epithelial-mesenchymal transition (EMT), drug resistance, autophagy, and the invasion, migration, apoptosis, and proliferation of OS cells are closely related to OS development (5-7). Unfortunately, the mechanism underlying OS progression has not been fully uncovered. Therefore, determining the key molecules implicated in OS may prove helpful to efforts to develop effective prevention and treatment measures.
Recently, a new series of noncoding RNAs (ncRNAs) have been found, including circular RNAs (circRNAs), long ncRNAs (lncRNAs, RNA transcripts >200 bp in length) and microRNAs (miRNAs, RNA transcripts ~22 bp in length), all of which have important effects on tumor development and the modulation of basic protein effectors of cellular functions (8,9). As the 2 main members of the ncRNA family, lncRNAs and miRNAs play pivotal roles in OS tumorigenesis.
An increasing bank of evidence has confirmed that numerous lncRNAs play key roles in multiple pathological and physiological cellular processes, including cell invasion, differentiation, apoptosis, and proliferation (10). Dysregulation of lncRNA expression has been found to have oncogenic effects (e.g., PROX1-AS1 in prostate cancer and NCK1-AS1 in urinary bladder cancer) (11,12) or tumor suppressive effects (e.g., TSLNC8 in breast cancer and RP11-422N16.3 in hepatocellular carcinoma) (13,14) during carcinogenesis. So far, a number of lncRNAs have been reported to possess promising prognostic or diagnostic value for OS (15,16); however, the role of prostate androgen-regulated transcript 1 (PART 1) in this malignancy is largely unknown. Recently, studies by showed that PART1 regulated the apoptosis of chondrocytes in osteoarthritis (17). A recent study by investigated the functions of PART1 in hepatocellular carcinoma and found that PART1 served as oncogenic lncRNA through sponging miR-590-3p to upregulate HMGB2 expression in hepatocellular carcinoma (18). Accordingly, we hypothesize that PART1 may play a key role in OS development.
In recent decades, miRNAs have also been found to serve as epigenetic regulators in disease development. MiRNAs repress gene expression and participate in gene silencing via direct interaction with the 3'-untranslated region (UTR) of target messenger RNAs (mRNAs), leading to the repression of mRNA translation or degradation (19). miRNAs participate in a variety of pathological and biological processes, including carcinogenesis, metabolism, and embryonic development (20). Several crucial activities of miRNAs in OS have been reported (21). Specifically, lncRNAs have been demonstrated to be endogenously competing RNAs which target miRNAs to inhibit miRNA-associated gene degradation.
In the present study, we investigated the expression and roles of PART 1 in OS, as well as the potential underlying regulatory mechanism. We elucidated that PART 1 serves as a competing endogenous RNA in OS by sponging miR-20b-5p. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-21-658).
Methods
Tissue samples
Forty-six pairs of OS tissue samples and matched non-cancerous tissues were harvested from patients who underwent excision surgery for OS in our hospital. All of the patients were radiation and chemotherapy naive. Liquid nitrogen was used to freeze the tissue samples before the extraction of total RNA. All patients signed a written informed consent form. All procedures in our study were carried out in accordance with the Helsinki Declaration (as revised in 2013). The study was approved by the Ethics Committee Board of our Hospital.
Cell lines and cell culture
OS cell lines [HOS (TCHu167) and MG-63 (TCHu124)] and human fetal osteoblastic cell line (hFOB) 1.19 were acquired from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS; Invitrogen) in a humidified chamber containing 5% CO2 at 37 °C.
Cell transfection
The miR-20b-5p inhibitor and mimic were designed by Gene Pharma (Shanghai, China). The whole sequences of PART 1 and bone morphogenic protein and activin membrane-bound inhibitor homolog (BAMBI) were cloned into pcDNA3.1 vector to overexpress PART 1 and BAMBI, respectively. For knockdown of PART 1, its small interfering RNAs (siRNAs) were synthesized as si-PART 1 by Gene Pharma (Shanghai, China). Lipofectamine 2000 (Invitrogen) was employed to transfect the above plasmids into HOS and MG-63 cells.
Real time-quantitative polymerase chain reaction (RT-qPCR) assay
TRIzol reagent (Invitrogen) was utilized for the extraction of total RNA from OS tissue samples and cultured cell lines, afterwards, a reverse transcription reaction was performed using a reverse transcription kit (Takara Bio Company, Shiga, Japan). qRT-PCR was completed with SYBR® Green PCR Master mix (Thermo Fisher Scientific, Inc., MA, USA) on an ABI Prism 7500 Sequence Detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.) in adherence to the manufacturers’ protocols and with U6 or GAPDH serving as an internal control. The 2−ΔΔCt method was used for measurement of relative gene expressions. The primers used were as follows: PART1 Forward, 5'-AAG GCC GTG TCA GAA CTC AA-3' and Reverse, 5'-GTT TTC CAT CTCA GCC TGG A-3'; miR-20b-5p forward, 5'-ACA CTC CAG CTG GGC AAA GTG CTC ATA GT-3' and reverse, 5'-TGG TGT CGT GGA GTC G-3'; BAMBI forward, 5'-CTC AAA TTC CCC ACT CAC CCA-3' and reverse, 5'-GCT GAT ACC TGT TTC CTT GTC CTG-3'; U6 forward, 5'-CTC GCT TCG GCA GCA CA-3' and reverse, 5'-AAC GCT TCA CGA ATT TGC GT-3'; GAPDH forward, 5'-AAT CCC ATC ACC ATC TTC CA-3' and reverse, 5'-TGG ACT CCA CGA CGT ACT CA-3'.
Cell Counting Kit-8 (CCK-8) assay
To determine cell viability, a CCK-8 assay was carried out as instructed by the manufacturer. OS cells were inserted into a 96-well plate and harvested at 24, 48, 72 or 96 hours post transfection. Then, after the indicated amount of time, CCK-8 was added to each well and the cells were incubated for a further 1 hour. The absorbance was detected at 450 nm using a microplate reader (Bio-Rad Laboratories, Hercules, CA, USA).
Transwell assay
The migration and invasion abilities of cells were detected by Transwell assay. Cell migration ability was determined using 6.5-mm Transwell chambers (8.0 µm pore size; BD Biosciences, Franklin Lakes, NJ, USA), and cell invasion ability was assessed using Transwell chambers precoated with Matrigel (BD Biosciences). Briefly, OS cells were resuspended in serum-free medium and then seeded into the apical chambers, with the bottom chambers filled with DMEM containing 10% FBS. After 24 hours of incubation, non-invasive or non-migratory cells were removed with cotton swabs. Cells located in the lower chamber were fixed and stained. Finally, cells in 5 randomly selected visual fields were quantified under a light microscope (Olympus Corp., Tokyo, Japan).
Western blot
Total protein extraction was accomplished using RIPA buffer (Beyotime, Shanghai, China). After measurement of the protein concentration using a bicinchoninic acid protein assay kit (Beyotime), the protein samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) separation and transferred onto a polyvinylidene difluoride (PVDF) membrane. The membrane was blocked with 5% skim milk and incubated with specific primary antibodies against BAMBI (ab203070; 1:1,000, Abcam, Cambridge, MA, USA) and GAPDH (ab9485; 1:2000, Abcam, Cambridge, MA, USA) at 4 °C. After incubation overnight, the membrane was incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit (1:2,000, Abcam, Cambridge, MA, USA) secondary antibody for 2 hours at room temperature. Finally, the signals were detected using an electrochemiluminescence (ECL) kit (Thermo Fisher Scientific, Inc.). GAPDH was used as the internal control.
Luciferase reporter assay
Dual-luciferase reporter assay (Promega, Madison, WI, USA) was performed to verify the relationships between PART1 or BAMBI and miR-20b-5p. After that, the wild-type PART1 and mutant PART1 sequences harboring predicted miR-20b-5p binding sites were synthesized and inserted into the pGL3-control vector (Promega, Madison, WI, USA) to construct the luciferase reporter vector of PART1-WT and PART1-Mut. Similarly, the wild-type BAMBI 3'-untranslated regions (UTR) and mutant BAMBI 3'-UTR sequences containing embracing predicted miR-20b-5p binding sites were synthesized and inserted into the pGL3-control vector for the construction of the luciferase reporter vectors of BAMBI-WT and BAMBI-Mut. Following this, the luciferase reporter vectors were cotransfected into OS cells with NC-mimics or miR-20b-5p mimics using Lipofectamine 2000 (Invitrogen) for the execution of the dual-luciferase reporter assay, respectively. Finally, cells were harvested in 48 hours post transfection and the luciferase activities of luciferase reporter vectors were evaluated via the dual-luciferase reporter assay kit (Promega).
Statistical analysis
All of the above experiments were performed in triplicate. SPSS 17.0 version (SPSS Inc., Chicago, IL, USA) was used to perform the statistical analyses. Data were tested using Student’s t-test or one-way analysis of variance with Tukey’s post-hoc test. The relationships between the expressions of miR-20b-5p and PART 1, PART 1 and BAMBI were assessed by Spearman’s or Pearson’s correlation analysis. The overall survival of the OS patients was determined with Kaplan-Meier curve together with log-rank test. P<0.05 was considered to indicate significant difference.
Results
High PART 1 expression in OS tissue indicated a poor prognosis
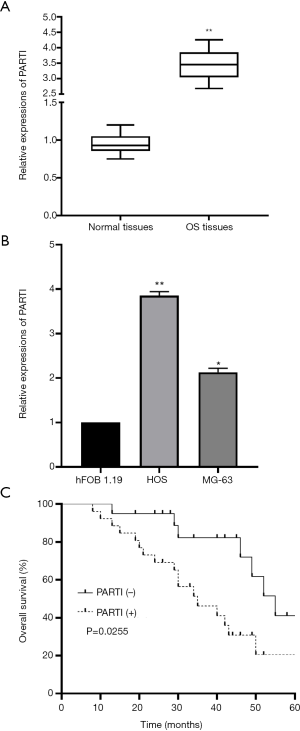
To determine the clinical significance of PART 1 in OS, we firstly detected the expression level of PART 1 in OS tissues and matched non-tumor tissues. The RT-qPCR results demonstrated that PART 1 expression was significantly increased in OS tissues compared to non-tumor tissues (Figure 1A). Similarly, upregulation of PART 1 was also observed in OS cells (Figure 1B). The survival analysis indicated that OS patients with PART 1 upregulation had strikingly shorter overall survival compared to the patients with lower PART 1 expression (Figure 1C). Overall, these results showed that PART 1 was upregulated in patients with OS and indicated a poor prognosis.
PART 1 accelerated OS cell proliferation, invasion, and migration
Having detected the aberrant up-regulation of PART 1 in OS tissues, we next performed functional assays, including a CCK-8 assay and Transwell assays, to determine the functions of PART 1 in the progression of OS. The cell lines MG-63 and HOS were transfected with pcDNA3.1-PART 1 or si-PART 1. The results of RT-qPCR verified that PART 1 was successfully overexpressed in MG-63 cells and was knocked down in HOS cells following transfection with pcDNA3.1-PART 1 or si-PART 1 (Figure 2A). The CCK-8 assay showed that pcDNA3.1-PART 1 significantly elevated the viability of MG-63 cells, whereas the proliferative ability of HOS cells was obviously reduced by si-PART 1 transfection (Figure 2A). Also, the Transwell assays revealed that PART 1 upregulation promoted the migration and invasion abilities of MG-63 cells (Figure 2B). In contrast, PART 1 knockdown notably reduced HOS cell migration and invasion (Figure 2B). Taken together, these observations suggested that PART 1 upregulation contributed to the malignant progression of OS.

PART 1 acted as a sponge of miR-20b-5p in OS cells
To determine the molecular mechanisms participating in PART 1-mediated OS progression, miRNAs could potentially serve as targets for PART 1 were predicted with Starbase. Results showed that PART 1 contained conserved binding sites for miR-20b-5p (Figure 3A). Subsequently, a luciferase reporter assay was performed to verify the correlation of PART 1 with miR-20b-5p. The miR-20b-5p mimics noticeably decreased the luciferase activities of the PART 1-wt plasmid; however, we failed to observe a notable difference in the luciferase activities of the PART 1-mut plasmid (Figure 3B). Next, the expression levels of miR-20b-5p in cells transfected with pcDNA3.1-PART 1 or si-PART 1 were measured by RT-qPCR. When PART 1 was overexpressed, miR-20b-5p expression was decreased, while PART 1 knockdown dramatically increased miR-20b-5p expression (Figure 3C). Similarly, the regulatory functions of miR-20b-5p in PART 1 expression were also investigated. As shown in Figure 3D, miR-20b-5p inhibition resulted in significant upregulation of PART 1, whereas the opposite effect was observed with miR-20b-5p overexpression. Additionally, in OS tissues, a significant decrease in miR-20b-5p expression was detected (Figure 3E), and a negative correlation between the expressions of PART 1 and miR-20b-5p was also confirmed (Figure 3F).

miR-20b-5p inhibited proliferation, invasion and migration in OS cells
The expression levels of miR-20b-5p in OS cells were further analyzed. A remarkable decrease in miR-20b-5p in OS cells was verified (Figure 4A). HOS and MG-63 cells were transfected with miR-20b-5p mimics or inhibitor to induce miR-20b-5p overexpression or inhibition, respectively. The transfection was confirmed to have been successfully completed by RT-qPCR (Figure 4B). The regulatory effects of miR-20b-5p on OS cell proliferation, invasion, and migration were subsequently investigated. The results showed that miR-20b-5p overexpression inhibited OS cell proliferation, invasion, and migration, while miR-20b-5p silencing exerted the opposite functions (Figure 4C,D).

BAMBI served as a target of miR-20b-5p in LAC cells
We further explored the mechanism underlying the promotion of OS progression by the PART 1/miR-20b-5p axis. TargetScan showed that BAMBI contained binding sites of miR-20b-5p (Figure 5A). The direct binding of miR-20b-5p to the 3'-UTR of BAMBI at putative sites was confirmed by the results of a luciferase reporter assay (Figure 5B). BAMBI was significantly inhibited by miR-20b-5p overexpression and promoted by miR-20b-5p inhibition (Figure 5C,D). Furthermore, BAMBI was markedly upregulated in OS tissue samples compared to the para-carcinoma tissues (Figure 5E). In addition, a positive correlation of the expressions of BAMBI and PART 1 was found to exist in OS tissues (Figure 5F).

PART 1 promoted OS tumorigenesis by sponging miR-20b-5p to upregulate BAMBI
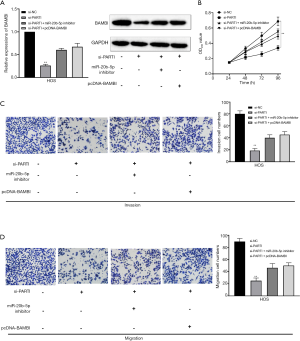
To determine whether the miR-20b-5p/BAMBI axis was implicated in the oncogenic functions of PART 1 in OS cells, a rescue assay was carried out. MiR-20b-5p inhibitor and pcDNA-BAMBI were transfected into OS cells together with si-PART 1. As shown in Figure 6A, BAMBI expression was significantly downregulated by si-PART 1, and this reduction was PART 1 ally reversed by silencing of miR-20b-5p or BAMBI overexpression. Additionally, we found that the suppressive effects of PART 1 knockdown on OS cell proliferation, migration, and invasion could be dramatically attenuated by miR-20b-5p silencing or BAMBI overexpression (Figure 6B,C,D). All of these data revealed PART 1 to be a modulator of OS cell malignancy through its sponging of miR-20b-5p to promote BAMBI expression.
Discussion
OS is a common bone malignancy in adolescents and teens. Tumor recurrence and metastasis are 2 primary factors for the high mortality rate of OS (22). There is mounting evidence showing that lncRNA is important in a variety of biological cellular processes (23,24). Previous studies have indicated that dysregulation of lncRNAs is correlated with tumor progression and patient outcomes (25). Recently, lncRNAs have been found to play crucial roles in OS. For instance, Ding et al. found that CRNDE (colorectal neoplasia differentially expressed) facilitated OS cell proliferation, invasion, and EMT via the Wnt/beta-catenin signaling pathway, following activation by SP1 (26). Furthermore, Cui et al. found that TMPO antisense RNA 1 promoted OS tumorigenesis by regulating the miR-199a-5p/WNT7B axis (27). Also, a study by Zhu et al. showed that PCAT6 promoted OS progression by sponging miR-185-5p and activating the transforming growth factor beta signaling pathway (28). Yet, the impact of PART 1 on the biological behavior of OS cells has remained unclear. Therefore, the aim of the present study was to elucidate the roles and mechanisms of PART 1 in OS.
LncRNA PART 1 is known as an androgen-regulated and prostate-specific gene (29). PART 1 overexpression in the prostate gland has been confirmed as being related to prostate tumor initiation (30). In recent years, aberrant PART 1 expression has also been confirmed in other tumors. For instance, Xuan et al.’s study indicated PART 1 was an independent predictor of prognosis in glioma patients (31), while Zhou et al. found that PART 1 regulated colorectal cancer via activation of the Wnt/beta-catenin pathway and regulation of miR-150-5p/miR-520h/CTNNB1 (32). Moreover, Zhu et al. found that PART 1 contributed to non-small cell lung cancer progression via the JAK-STAT signaling pathway (33). In our study, we found that PART 1 was upregulated in OS, with its overexpression promoting the viability, invasion, and migration of OS cells.
Accumulating studies have demonstrated that lncRNAs may serve as ceRNAs in carcinogenesis. ceRNAs can sponge miRNAs, reducing their binding to the target genes and thereby modulating their expression (34). In our study, miR-20b-5p, which was predicted as a target of PART 1, was found at low levels in OS tissues and cells. Further investigation of the potential mechanisms indicated that BAMBI served as a direct target of miR-20b-5p in OS cells. PART 1 was found to act as a promoter of OS tumorigenesis by sponging miR-20b-5p to upregulate BAMBI.
In conclusion, the data of the present study revealed that high levels of PART 1 and low levels of miR-20b-5p are expressed in OS. PART 1 upregulation notably contributed to OS cell proliferation and cell mobility. Further study showed that the anti-OS functions of PART 1 were exerted via its sponging of miR-20b-5p to up-regulate BAMBI. Our findings may provide novel diagnostic markers for OS and enrich our knowledge of OS progression.
Acknowledgments
Funding: This work was supported by grants from the National Natural Science Foundation of China (81702671), Jiangxi Provincial Natural Science Foundation (20202BABL216047, 20171BAB205031), and the Department of Science and Technology of Jiangxi Province (S2019ZPYFB2011).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-658
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-658
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-658). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All patients signed a written informed consent form. All procedures in our study were carried out in accordance with the Helsinki Declaration (as revised in 2013). The study was approved by the Ethics Committee Board of our Hospital.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Oh JY, Kim EH, Lee YJ, et al. Synergistic Autophagy Effect of miR-212-3p in Zoledronic Acid-Treated In Vitro and Orthotopic In Vivo Models and in Patient-Derived Osteosarcoma Cells. Cancers (Basel) 2019;11:1812. [Crossref] [PubMed]
- Friebele JC, Peck J, Pan X, et al. Osteosarcoma: A Meta-Analysis and Review of the Literature. Am J Orthop (Belle Mead NJ) 2015;44:547-53. [PubMed]
- Jiang Y, Wang X, Cheng Y, et al. Associations between inflammatory gene polymorphisms (TNF-alpha 308G/A, TNF-alpha 238G/A, TNF-beta 252A/G, TGF-beta1 29T/C, IL-6 174G/C and IL-10 1082A/G) and susceptibility to osteosarcoma: a meta-analysis and literature review. Oncotarget 2017;8:97571-83. [Crossref] [PubMed]
- Qi L, Ren X, Liu Z, et al. Predictors and Survival of Patients with Osteosarcoma After Limb Salvage versus Amputation: A Population-Based Analysis with Propensity Score Matching. World J Surg 2020;44:2201-10. [Crossref] [PubMed]
- Zhang Y, Wang F, Wang L, et al. MiR-363 suppresses cell migration, invasion, and epithelial-mesenchymal transition of osteosarcoma by binding to NOB1. World J Surg Oncol 2020;18:83. [Crossref] [PubMed]
- Yu WX, Lu C, Wang B, et al. Effects of rapamycin on osteosarcoma cell proliferation and apoptosis by inducing autophagy. Eur Rev Med Pharmacol Sci 2020;24:915-21. [PubMed]
- Zhang Y, Weng Q, Chen J, et al. Morusin inhibited human osteosarcoma via PI3K-AKT signaling pathway. Curr Pharm Biotechnol 2020;21:1402-9. [Crossref] [PubMed]
- Zhou Y, Li X, Yang H. LINC00612 functions as a ceRNA for miR-214-5p to promote the proliferation and invasion of osteosarcoma in vitro and in vivo. Exp Cell Res 2020;392:112012 [Crossref] [PubMed]
- Luo M, Liang C. LncRNA LINC00483 promotes gastric cancer development through regulating MAPK1 expression by sponging miR-490-3p. Biol Res 2020;53:14. [Crossref] [PubMed]
- Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer 2018;18:5-18. [Crossref] [PubMed]
- Qian C, Liao CH, Tan BF, et al. LncRNA PROX1-AS1 promotes proliferation, invasion, and migration in prostate cancer via targeting miR-647. Eur Rev Med Pharmacol Sci 2020;24:2938-44. [PubMed]
- Qiao Z, Dai H, Zhang Y, et al. LncRNA NCK1-AS1 Promotes Cancer Cell Proliferation and Increase Cell Stemness in Urinary Bladder Cancer Patients by Downregulating miR-143. Cancer Manag Res 2020;12:1661-8. [Crossref] [PubMed]
- Qin CX, Yang XQ, Jin GC, et al. LncRNA TSLNC8 inhibits proliferation of breast cancer cell through the miR-214-3p/FOXP2 axis. Eur Rev Med Pharmacol Sci 2019;23:8440-8. [PubMed]
- Sun Y, Zhou Q, Li J, et al. LncRNA RP11-422N16.3 Inhibits Cell Proliferation and EMT, and Induces Apoptosis in Hepatocellular Carcinoma Cells by Sponging miR-23b-3p. Onco Targets Ther 2019;12:10943-61. [Crossref] [PubMed]
- Misawa A, Orimo H. lncRNA HOTAIR Inhibits Mineralization in Osteoblastic Osteosarcoma Cells by Epigenetically Repressing ALPL. Calcif Tissue Int 2018;103:422-30. [Crossref] [PubMed]
- Zhang N, Meng X, Mei L, et al. LncRNA DLX6-AS1 promotes tumor proliferation and metastasis in osteosarcoma through modulating miR-641/HOXA9 signaling pathway. J Cell Biochem 2019; Epub ahead of print. [Crossref] [PubMed]
- Lu C, Li Z, Hu S, et al. LncRNA PART-1 targets TGFBR2/Smad3 to regulate cell viability and apoptosis of chondrocytes via acting as miR-590-3p sponge in osteoarthritis. J Cell Mol Med 2019;23:8196-205. [Crossref] [PubMed]
- Pu J, Tan C, Shao Z, et al. Long Noncoding RNA PART1 Promotes Hepatocellular Carcinoma Progression via Targeting miR-590-3p/HMGB2 Axis. Onco Targets Ther 2020;13:9203-11. [Crossref] [PubMed]
- Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis 2012;33:1126-33. [Crossref] [PubMed]
- Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development 2005;132:4653-62. [Crossref] [PubMed]
- Patil SL, Palat A, Pan Y, et al. MicroRNA-509-3p inhibits cellular migration, invasion, and proliferation, and sensitizes osteosarcoma to cisplatin. Sci Rep 2019;9:19089. [Crossref] [PubMed]
- Basile P, Greengard E, Weigel B, et al. Prognostic Factors for Development of Subsequent Metastases in Localized Osteosarcoma: A Systematic Review and Identification of Literature Gaps. Sarcoma 2020;2020:7431549 [Crossref] [PubMed]
- Kawasaki Y, Miyamoto M, Oda T, et al. The novel lncRNA CALIC upregulates AXL to promote colon cancer metastasis. EMBO Rep 2019;20:e47052 [Crossref] [PubMed]
- Carlevaro-Fita J, Lanzos A, Feuerbach L, et al. Cancer LncRNA Census reveals evidence for deep functional conservation of long noncoding RNAs in tumorigenesis. Commun Biol 2020;3:56. [Crossref] [PubMed]
- Wang W, Xie Y, Chen F, et al. LncRNA MEG3 acts a biomarker and regulates cell functions by targeting ADAR1 in colorectal cancer. World J Gastroenterol 2019;25:3972-84. [Crossref] [PubMed]
- Ding Q, Mo F, Cai X, et al. LncRNA CRNDE is activated by SP1 and promotes osteosarcoma proliferation, invasion, and epithelial-mesenchymal transition via Wnt/beta-catenin signaling pathway. J Cell Biochem 2020;121:3358-71. [Crossref] [PubMed]
- Cui H, Zhao J. LncRNA TMPO-AS1 serves as a ceRNA to promote osteosarcoma tumorigenesis by regulating miR-199a-5p/WNT7B axis. J Cell Biochem 2020;121:2284-93. [Crossref] [PubMed]
- Zhu C, Huang L, Xu F, et al. LncRNA PCAT6 promotes tumor progression in osteosarcoma via activation of TGF-beta pathway by sponging miR-185-5p. Biochem Biophys Res Commun 2020;521:463-70. [Crossref] [PubMed]
- Sidiropoulos M, Chang A, Jung K, et al. Expression and regulation of prostate androgen regulated transcript-1 (PART-1) and identification of differential expression in prostatic cancer. Br J Cancer 2001;85:393-7. [Crossref] [PubMed]
- Lin B, White JT, Ferguson C, et al. PART-1: a novel human prostate-specific, androgen-regulated gene that maps to chromosome 5q12. Cancer Res 2000;60:858-63. [PubMed]
- Xuan C, Jin M, Wang L, et al. PART1 and hsa-miR-429-Mediated SHCBP1 Expression Is an Independent Predictor of Poor Prognosis in Glioma Patients. Biomed Res Int 2020;2020:1767056 [Crossref] [PubMed]
- Zhou T, Wu L, Ma N, et al. LncRNA PART1 regulates colorectal cancer via targeting miR-150-5p/miR-520h/CTNNB1 and activating Wnt/beta-catenin pathway. Int J Biochem Cell Biol 2020;118:105637 [Crossref] [PubMed]
- Zhu D, Yu Y, Wang W, et al. Long noncoding RNA PART1 promotes progression of non-small cell lung cancer cells via JAK-STAT signaling pathway. Cancer Med 2019;8:6064-81. [Crossref] [PubMed]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014;505:344-52. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)


