
High-throughput sequencing profile of laryngeal cancers: analysis of co-expression and competing endogenous RNA networks of circular RNAs, long non-coding RNAs, and messenger RNAs
Introduction
Laryngeal cancer (LC) represents approximately 30–50% of all neoplasms of the head and neck, with 177,422 new cases and 94,771 deaths worldwide in 2018 (1). During the past 40 years, the 5-year survival rate of LC has declined from 66% to 63%, presenting a negative trend (2). The initiation and development of LC is a complicated process caused by a variety of pathogenic factors and multiple genetic disorders (g). LC may result from a series of genetic changes caused by smoking, drinking, papillomavirus infection, or exposure to harmful dust (3,4). As for genes dysregulated genes, an increasing number of studies have shown that the dysregulation of multiple genes leads to the malignant progression of many tumors, especially LC (5). For instance, LncRNA MEG3 inhibits cell proliferation and induces apoptosis in laryngeal cancer via miR-23a/APAF-1 axis (6). However, the specific pathogenesis of LC remains unclear, and there is a lack of ideal biomarkers for diagnosis, treatment, and prognosis.
To further study the molecular mechanisms of LC tumorigenesis and progression, we sequenced 5 pairs of LC tissues. Human genome sequencing analysis showed that only 2% of DNA with coding function accounted for the whole genome sequence. Therefore, most of the transcription products of these DNA sequences were non-coding RNAs. With the development of high-throughput sequencing and bioinformatics, many new non-coding RNAs (ncRNA) have been discovered. For decades, ncRNAs were considered meaningless transcriptional noise. However, ncRNAs have now been confirmed to be involved in the tumorigenesis and progression of many diseases, including tumors (7,8). Long non-coding RNAs (lncRNAs) are a kind of ncRNA with lengths of more than 200 nucleotides, and play important roles at both the transcriptional and post-transcriptional level (9). Circular RNAs (circRNAs) have attracted much attention during recent years. Unlike traditional linear RNA, circRNAs area new type of ncRNA characterized by a single-stranded closed-loop structure formed by 3’-5’ terminal reverse splicing of mRNA precursor (pre-mRNA) (10). CircRNAs have the following defining characteristics: tolerance to RNA exonuclease digestion, strong stability, high expression abundance, and high tissue specificity. Therefore, they are likely to be ideal biomarkers for some diseases, especially tumors (11,12). In recent years, it has been reported that ncRNAs participate in the mechanism of competing endogenous RNAs (ceRNAs). These are circRNAs or lncRNAs that act as sponges for microRNA (miRNA). Once they have absorbed specific miRNAs, ceRNAs scan then regulate the expression of coding genes (13). Studies have shown that circRNAs and lncRNAs play an important role in the progression of LC (14,15), but no sensitive, widely used, and reliable indicators for the diagnosis or prediction of LC have been found. Therefore, further studies on circRNAs and lncRNAs will be helpful for the diagnosis, treatment, and prognosis of LC.
Due to the development of second-generation high-throughput sequencing technology, a large number of ncRNAs have been identified in various species, cells, and clinical samples. However, most circRNAs and lncRNAs remain functionally undefined and lack sufficient annotation. Currently, there is a gap in the data regarding transcriptome expression profile analysis based on high-throughput sequencing in LC. The molecular regulation mechanisms of circRNAs and lncRNAs in LC are still not well understood and require further study. Hence, in order to clarify the functions and mechanisms of ncRNAs in LC, we paired fresh-frozen LC tissues with noncancerous tissues from 5 selected patients, and sequenced them to study differentially expressed circRNAs, lncRNA, and mRNAs. Through comparison using the circBase and RefSeq databases, the splicing sequence composition of the predicted sequence was confirmed. Differentially expressed ncRNAs were screened and identified. Furthermore, RT-qPCR was used to validate the representative circRNAs, lncRNAs, and mRNAs. We also carried out co-expression network and ceRNA network analyses, and conducted pathway enrichment analysis using the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. These analyses were performed on significantly dysregulated expressed genes to identify the functions and mechanisms of differentially expressed genes.
Recently, accumulating studies have investigated the molecular mechanisms of LC (16). And several studies have demonstrated that noncoding RNAs can play crucial roles in the pathogenesis of LC (14,17). However, different from those studies on LC, our study is conducive to the discovery of new LC transcripts, which allows for the analysis and identification of differentially expressed ncRNAs in patients with LC. Also, we proposed a novel lncRNA with ideal prognostic value, and predicted a possible target pathway of this lncRNA. Thus, our research provides an overview of the expression of ncRNAs in LC, as well as a scientific basis for the discovery of new biomarkers for the disease. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-21-584).
Methods
Patients and samples
All specimens were obtained with the informed, written consent of patients and their families, and were approved by the ethics committee of the First Affiliated Hospital of China Medical University. We collected 47 pairs of LC and noncancerous tissues from patients who underwent total or partial laryngectomy. In vitro specimens were immediately washed repeatedly with frozen phosphate-buffered saline (PBS) 3 times, quickly collected, and stored in liquid nitrogen, then transferred to a −80 °C freezer for preservation. According to the LC staging criteria stipulated by the World Health Organization (WHO), pathological diagnosis and classification of specimens were completed by 2 professional pathologists. Inclusion criteria: (I) clear pathological diagnosis of laryngeal cancer; (II) new cases of LC; (III) the patient did not receive any radiotherapy, chemotherapy or other anticancer treatments before surgery. Exclusion criteria: (I) the diagnosis was not clear, such as atypical hyperplasia, laryngeal keratosis and other non-cancerous lesions; (II) recurrent cases; (III) cases receiving treatment other than surgery. The study was approved by the Ethics Committee of the First Affiliated Hospital of China Medical University (number [2016]2016-30-2), and informed consent was obtained from all patients. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture
The human LC cell lines LCC, LLN, and TU212 were obtained from the Institute of Basic Medical Science, Chinese Academy of Medical Sciences & School of Basic Medicine, Peking Union Medical College. The normal cell line HaCaT was obtained from the LC Research Office (the First Affiliated Hospital of China Medical University, Shenyang, China). All cell lines were maintained at 37 °C in RPMI 1640 medium (Gibco, USA) with 10% fetal bovine serum (FBS) (Bovogen, Australia), and were incubated in a humidified atmosphere containing 5% CO2. All cell lines were authenticated by STR DNA profiling and tested for Mycoplasma (GENEWIZ Co, Ltd, Suzhou, China) within 6 months.
RNA extraction
Trizol (Invitrogen, USA) was used to extract nucleic acids from both cancerous and noncancerous tissues according to the manufacturer’s instructions. The RNA purity and concentration were assessed by optical density (OD)at A260/280 (>1.8) and A260/A230 (>1.6). RNA quality and integrity were checked using a 2100 Bioanalyzer (Agilent Technologies, USA) and the RNA integrity number (RIN) was found to be >7.0.
Sequencing analysis
Five out of 47 pairs of specimens were selected for sequencing. The preparation of whole transcriptome libraries and deep sequencing were conducted by Hua Da Gene Corp., Ltd. (Shenzhen, China). The whole transcriptome libraries were established using the New England Biolabs (NEB) Next Ultra Directional RNA Library Prep Kit for Illumina according to the manufacturer’s instructions. Libraries were tested for quality using the Agilent 2100 Bioanalyzer system and the StepOnePlus RT-PCR (Applied Biosystems, USA) system. The qualified libraries were sequenced on an Illumina HiSeq X Ten instrument in paired-end form, generating 150 nucleotides. Clean reads were aligned to the human genome (GRCh38) using the HISAT program (18) and alignment results were reconstructed with cuff compare. The NONCODE and Pfam databases were used as annotation references for lncRNA and mRNA analyses. INFERNAL (19) was used to predict lncRNAs. CIRI (20) and find_circ (10) were used to predict circRNAs. The read count of each transcript was normalized in the form of fragments per kilobase of exon per million mapped reads (FPKM). In our study, differentially expressed genes and transcripts were identified using the following criteria: |log2(fold-change)| ≥1 and P value ≤0.05 between the two samples.
Quantitative real-time PCR validation
Total RNA was isolated from the 47 pairs of LC and noncancerous tissues, then was reverse transcribed to cDNA using M-MLV reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. Subsequently, qRT-PCR was conducted in 3 independent wells with a total reaction volume of 20 µL. This included10 µL of SYBR Green Master Mix, 2 µL of cDNA, 0.4 µL of ROX, 0.8 µL of PCR Forward Primer (10 µM), 0.8 µL of PCR Reverse Primer (10 µM), and 6 µL of nuclease-free water. β-actin was used as a reference. The program was set at 95 °C for 5 min, then at 95 °C for 5 sec and 60 °C for 34 s, for a total of 40 cycles. The RNA expression levels were assessed by the 2−ΔΔCt method (21). Student’s t-tests were used with a standard of P<0.05. Data are presented as mean ± SD. The primer sequences are shown in Table S1.
Western blotting
Radioimmunoprecipitation buffer (Beyotime, Beijing, China) was used for lysing cellson ice within 30 min. The bicinchoninic acid (Beyotime, Beijing, China) was used for quantifying the protein concentrations. Lysates were resolved by SDS-PAGE and transferred onto polyvinylidene fluoride membranes (Millipore, Bedford, MA, USA). The membranes were blocked and incubated with primary antibodies against N-cadherin (ab76011, 1:1,000, Abcam, Cambridge, MA, USA), E-cadherin (ab212059, 1:1,000, Abcam, Cambridge, MA, USA), β-catenin (ab223075, 1:1,000, Abcam, Cambridge, MA, USA) and snail (ab216347, 1:1,000, Abcam, Cambridge, MA, USA) overnight at 4 °C and then incubated with HRP-conjugated secondary monoclonal antibody (ab205718, 1:5,000, Abcam, Cambridge, MA, USA) for 1 h at room temperature. The chemiluminescence system (Bio-Rad) was used for quantifying the protein of the gels.
RNA fluorescence in situ hybridization (RNA-FISH)
RNA-FISH analysis in LC cells was carried out according to the standard protocol. The cells were seeded on coverslips and fixed with 4% paraformaldehyde for 20 min. The specific probe sequence of SNHG29 for FISH was as follows: 5'-CCAGCTTAAAGAACTTTGCCCACCAACATAACCAA-3'. Cell nuclei were stained with DAPI (Thermo Fisher, USA). The images were photographed using a Nikon inverted fluorescence microscope. The fluorescence intensity was analyzed using ImageJ software (Maryland, USA).
Co-expression analysis
To calculate the correlation of expression levels between dysregulated circRNAs or lncRNAs and protein-coding mRNAs, circRNA-mRNA and lncRNA-mRNA co-expression networks were generated according to Pearson’s correlation coefficient. RNAs which were collected for further analysis had to show a coefficient parameter value >0.99 or <−0.99 with P<0.05.
CeRNA network analysis
A ceRNA network was formed by circRNAs, lncRNAs, miRNAs, and corresponding mRNAs. MiRanda (www.microrna.org) was used to predict the possible target miRNAs of dysregulated circRNAs or lncRNAs. Cytoscape (www.cytoscape.org) was used to generate a ceRNA network.
GO and KEGG pathway analysis
GO analysis (http://www.geneontology.org) was used to understand gene function. KEGG pathway analysis was used to study the mechanisms and interactions among differentially expressed genes.
Statistical analysis
Statistical significance was determined using Student’s t-test, with SPSS 22.0 (IBM, Armonk, NY, USA). MeV4 (http://mev.tm4.org) and GraphPad Prism 6 (GraphPad, San Diego, USA) were used to generate figures. P<0.05 was considered to indicate statistical significance, and P<0.01 was considered to indicate high significance.
Results
Results of sequencing
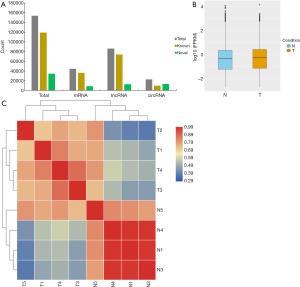
There were 1,594.81 million raw reads in the original sequencing data. After filtration, 1,566.87 million (98.2%) clean reads remained for the further analysis, and 84.86% of clean reads were mapped to the human reference genome. Finally, a total of 119,363 known transcripts and 34,313 novel transcripts were obtained (Figure 1A). Furthermore, we identified 62,163 genes in cancer tissue and 66705 genes in noncancerous tissue. Approximately 1.3% of the expression of all transcripts was above 50 FPKM, with nearly 49.6% lower than 0.5 FPKM (Figure 1B). The heatmap was drawn to evaluate the inter-sample correlation on the basis of the Pearson’s correlation coefficient of the transcript expression levels (Figure 1C). A discrepancy was apparent between the cancer group and the normal group, but only slightly within the intra-group. Our data quality was credible enough for further study.
Differentially expressed genes and transcripts
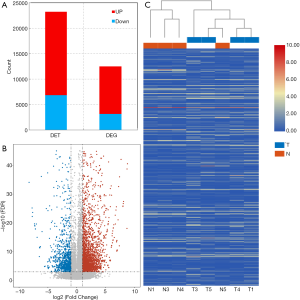
Overall, 43,320 significantly differentially expressed transcripts were identified, with a fold change ≥2.0 (P<0.05, adjusted P<0.1). Of these, 31,763 transcripts were upregulated, and 11,557 were downregulated (Figure 2A). The significant differences of transcript expression levels between cancerous and normal tissues were determined using volcano plot (Figure 2B) and clustering analysis (Figure 2C). The differentially expressed transcripts qualified for further analysis in the study of the molecular mechanisms of LC.
Construction of the co-expression network
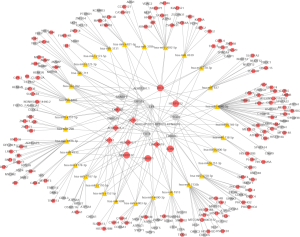
Since ncRNAs have no protein-coding capabilities, their functions can be predicted by protein-coding mRNAs. Therefore, co-expression analysis based on the expression of dysregulated transcripts was conducted to predict the functions of these ncRNAs. We selected significantly differentially expressed transcripts to generate a ncRNAs-mRNAs co-expression network (Figure 3 and https://cdn.amegroups.cn/static/public/10.21037atm-21-584-1.pdf). EPB41L4B, which may promote the invasive ability of cancer cells and indicate poor prognosis (22), was correlated with the expression of ADRM1 and SNHG29. STAT6, which is correlated with cancer cell growth (23), was co-expressed with PALLD, WFDC21P, and ADRM1.Through this network, we can predict the functions of ncRNAs based on protein-coding mRNAs.

Construction of the ceRNA network
Literature has shown that a multitude of ncRNAs can compete for binding to various miRNAs because of the similar sequence fragments (24,25). Therefore, a ceRNA network which mapped the correlations among circRNAs, lncRNAs, miRNAs, and mRNAs was constructed. Ten significantly differentially expressed circRNAs and lncRNAs were brought into the network (Figure 4 and https://cdn.amegroups.cn/static/public/10.21037atm-21-584-2.pdf). It has been reported that ZNF146 is upregulated in colorectal cancer and is involved in the progression of the disease (26). In our study, TBCD was found to harbor hsa-miR-3692-5p, affecting the expression of ZNF146. Therefore, ceRNA networks may help us better understand the mechanisms of ncRNAs in LC.
GO and KEGG analysis
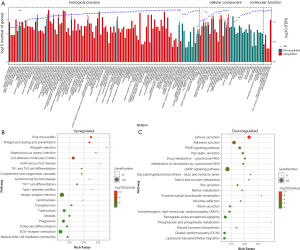
GO and KEGG analyses were performed to better understand the mechanisms of LC tumorigenesis. The terms which were most significantly enriched by GO analysis are shown in Figure 5A and https://cdn.amegroups.cn/static/public/10.21037atm-21-584-3.xlsx. The biological processes relating to the most significantly upregulated genes were biological regulation, positive regulation of cell-cell adhesion, and cell migration. Cell adhesion was found to be associated with genes which were clearly downregulated. This may relate to LC tumorigenesis. The most obviously upregulated and downregulated KEGG pathways are shown in Figure 5B,C, https://cdn.amegroups.cn/static/public/10.21037atm-21-584-4.xlsx, and https://cdn.amegroups.cn/static/public/10.21037atm-21-584-5.xlsx. The pathways which were found to be significantly upregulated involved cell adhesion molecules (CAMs) and antigen processing and presentation. The most downregulated pathways were those relating to adherens junction and peroxisome proliferator-activated receptor (PPAR) signaling. GO and KEGG analyses indicated that aberrant cell migration or adhesion maybe involved in the pathogenesis of LC.
Confirmation of transcripts and identification of novel ncRNAs
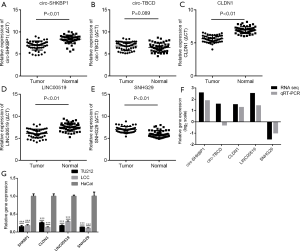
From the significantly differentially expressed circRNAs, lncRNAs, and mRNAs, we selected 2 circRNAs, 2 lncRNAs, and 1 mRNA through databases and literature analysis. We identified the transcripts as novel RNAs when the transcripts did not align to the circBase or RefSeq databases. Subsequently, qRT-PCR was performed on 47 pairs of tissue samples to verify the selected transcripts. The expression of all the selected transcripts except circ-TBCD was in accordance with the results of the sequencing data. The results are shown in Figure 6A,B,C,D,E,F and Table 1. Next, RT-qPCR was used to evaluate the expression of SKBP1, CLDN1, LINC00519, and SNHG29 at the cellular level (Figure 6G). The expression trend of SNHG29 in cell lines was finally confirmed to be in accordance with that in tissues. Therefore, we chose SNHG29 as our target gene for further study.

Full table
The characteristics and anticancer role of SNHG29
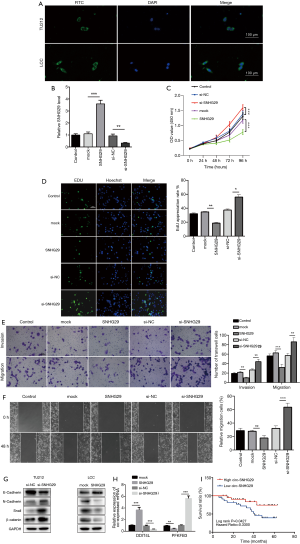
Next, we confirmed the characterization of SNHG29. FISH analysis was performed to validate the localization of SNHG29, which indicated that SNHG29 was localized in the nucleus and cytoplasm of TU212 and LCC cells (Figure 7A), but mainly localized in the cytoplasm. Previous results showed that SNHG29 had lower expression in the 2 LC cell lines compared with the normal control line HaCat (Figure 6G). To confirm the biological functions of SNHG29 in LC, we performed gain- or loss-of-function studies by transfection with SNHG29 or si-RNA, respectively. RT-qPCR was performed to validate the transfection efficiency (Figure 7B). Furthermore, we investigated the effects of SNHG29 on LC proliferation, migration, and invasion using related assays. Overexpression of SNHG29 markedly inhibited cell proliferation, whereas knockdown of SNHG29 significantly enhanced cell viability (Figure 7C). Subsequently, we performed the EDU assay to analyze the effects of SNHG29 on LC cells, and found that higher SNHG29 inhibited cell proliferation, whereas cell viability was promoted by overexpressed SNHG29 (Figure 7D). Additionally, the Transwell assay was performed in LCC cells transfected with si-RNA or SNHG29, which showed that more migrated cells were found in the membrane of wells inoculated with LC cells transfected with si-RNA, whereas fewer migrated cells were observed in the SNHG29 group (Figure 7E). Furthermore, overexpression of SNHG29 significantly inhibited the wound healing ability of LC cells. Conversely, downregulation of SNHG29 significantly enhanced cell motility (Figure 7F). Our results confirmed the role of SNHG29 in inhibiting LC cell growth. Importantly, the western bolt assay was used to observe the effects of SNHG29 having on the epithelial to mesenchymal transition process, indicating that SNHG29 upregulation inhibited the epithelial to mesenchymal transition process, whereas SNHG29 downregulation promoted the epithelial to mesenchymal transition process. The results indicated that SNHG29 inhibited the metastasis of laryngeal cancer (Figure 7G). Consequently, we overexpressed or knocked down SNHG29, and RT-qPCR was conducted to validate the expression levels of the co-expressed mRNAs of SNHG29. Results showed that 2 out of 13 mRNAs were altered following SNHG29 transfection, namely DDIT4L and PFKFB3 (Figure 7H). Importantly, we found that upregulation of SNHG29 was related to the higher survival rates (hazard ratio =0.3300, P=0.0427) (Figure 7I).
Correlation between SNHG29 level and clinical parameters
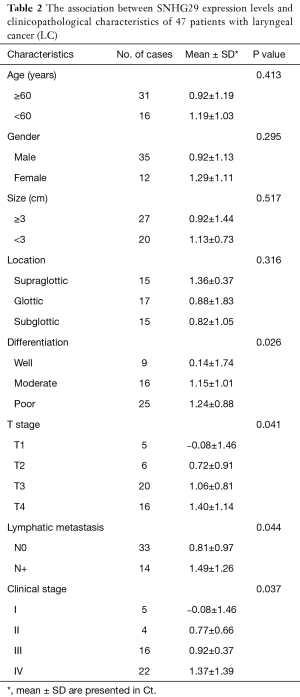
We investigated the relationship between SNHG29 expression and the clinicopathological parameters of the included patients (Table 2). Downregulation of SNHG29 was significantly associated with differentiation (P=0.026), T-stage (P=0.041), lymphatic metastasis (P=0.044), and clinical stage (P=0.037), but not with age, gender, tumor size, or tumor location.
Full table
Discussion
The genesis and progression of LC are comprehensive results of multiple factors, such as dysregulation of genes and pathological factors. At the molecular scale, LC may result from the abnormalities of multiple DNAs, RNAs and proteins (27,28). As new classes of noncoding RNAs, circRNAs and lncRNAs were found to play crucial roles in LC development. For example, lncRNA SOX2-OT was found to regulate laryngeal cancer cell proliferation, migration and invasion and induces apoptosis by suppressing miR-654 (29). Besides smoking, drinking and air pollution, high-risk HPV (HR-HPV) infection was found to be one of the most important pathogenic factors of LC (30). Interestingly, HPV infection affects the LC tumorigenesis via genomic integration, activating proto-oncogenes and inhibiting anti-oncogenes, indicating that the genetic dysregulation and pathological factors may interplay each other and both promote the LC development (31). Emerging studies indicated that alterations of microbiota structure may contribute to the initiation of LC (32). For instance, a higher α-diversity was found in cancer patients compared with control subjects and normal tissues adjacent to the tumor sites, as well as the relative abundances of seven bacterial genera differed among the three groups of samples. Interestingly, relative evidence showed that gene mutation elevated the risk of laryngeal cancer, indicating that mutation profile analysis may be used to determine laryngeal dysplasia at risk of developing invasive cancer (33). However, the underlying mechanism remains largely unknown. Thus, our investigation of genes abnormal alterations will contribute to the better understanding the pathogenesis mechanisms of LC.
The second generation of high-throughput sequencing is a rapidly developing and emerging technology characterized by high efficiency, high-throughput, high coverage, and high sensitivity. It relies on the use of the correct amount of interruption reagent in order to fragment the RNA, disrupt it, and use it as the template for reverse transcription. Specific binding ligands are subsequently added at both ends of the sequence, which is then ready for bridge PCR amplification. Finally, the constructed cDNA library is then purified and sequenced (34). Although the prediction and identification of transcriptomes based on high-throughput sequencing has been quite extensive (35), the functions and mechanisms of most ncRNAs are still unclear due to the limitations of current detection and identification techniques. The exploration and identification of new ncRNAs will be of great significance for determining the mechanisms of tumorigenesis and progression. Due to its high incidence and metastasis rate, LC could be a valuable choice for further study. Therefore, we performed the first transcriptome high-throughput sequencing of LC specimens. These specimens contained high and medium differentiated squamous cell carcinomas without metastasis. We found many new transcripts with differential expression, which are bound to play crucial roles in the occurrence and development of LC.
To clarify the reliability and application value of the screening results, we selected certain screening genes which displayed equal expression trends and confirmed their expression using qRT-PCR. Results showed that the circRNAs SHKBP1 and TBCD, the lncRNAs LINC000519 and SNHG29, and mRNA CLDN1 were amplified in qRT-PCR. The expression trend of qRT-PCR was in accordance with most sequencing results, except for that of circ-TBCD. By means of qRT-PCR and high-throughput sequencing, the selected RNAs, except for circ-TBCD, were confirmed as significantly differentially expressed between cancerous tissues and the control group. However, when we validated the expression levels in cell lines with RT-qPCR, only SNHG29 was the same among the sequencing data, tissue RT-qPCR, and cell RT-qPCR. Consequently, relative biological functional assays were conducted to illustrate that SNHG29 plays crucial roles in the tumorigenesis and progression of LC.
Most of our sequencing data was found to be in accordance with the existing literature. For example, it has been reported that WFDC21P showed low expression in hepatocellular carcinoma, related to the proliferation and metastasis of this tumor (36). Consistent with this study, our sequencing results showed that WFDC21P was also significantly downregulated in LC. Furthermore, MutL homolog 1 (MLH1), which was shown to be correlated with the prognosis of LC, was found to be downregulated in LC tissues (37). Similarly, our sequencing results showed that MLH1 had lower expression, indicating that MLH1 may be involved in the tumorigenesis of LC. The evidence above indicated that our data was reliable for further study.
Recent studies have found that changes in expression of ncRNAs have a certain correlation with their parent genes, and some ncRNAs derived from parent genes are also highly expressed in tumors and have functions similar to those of their parent genes (38,39). Therefore, by plotting the co-expression analysis network, we annotated and predicted the functions of ncRNAs according to the functions of mRNAs. To obtain a more reliable regulatory relationship, we selected tumor-related mRNAs to predict the functions of ncRNAs and set the Pearson’s correlation coefficient limit to be less than −0.99 or greater than 0.99 to draw a visual co-expression network. For example, NFIL3 was reported to promote lung cancer cell migration and is associated with lung cancer invasiveness (40). This corresponded to the co-expressed circRNAPPFIA1 and PCNXL2, which may be involved in promoting LC invasion and metastasis. CELF6, which is reported to be a potential suppressor, and is involved in cell proliferation and cell cycle progression via the modulation of p21 stability (41), was upregulated in our sequencing data as well as in the GEPIA database. The co-expressed circRNAs TBC1D31, GLS, and BPTF may also be involved in the tumorigenesis and development of LC. In our results, among the co-expressed mRNAs with SNHG29, DDIT4L and PFKFB3 were found to have altered expression when SNHG29 was overexpressed or silenced. DDIT4L and PFKFN3 have been found to be related to autophagy, and play crucial roles in cancers (42,43). Our assay validated that SNHG29 plays anticancer roles in LC. Through co-expression analysis, we could predict the functions and mechanisms of the co-expressed circRNAs and lncRNAs. Therefore, this analysis may prove to be an ideal method for researching ncRNAs.
At present, the mechanisms of ncRNAs mainly focus on ceRNA mechanisms. NcRNAs play a role by absorbing miRNAs in a “sponge-like” manner, and by inhibiting miRNAs, thereby affecting the expression of downstream mRNAs. For example, it has been reported that the high expression of circAGFG1 can block the inhibition of mir-195-5p on its target gene (CCNE1), thus promoting the invasion and metastasis of breast cancer (44). Therefore, we screened out the circRNAs and lncRNAs with large differences from the results of high-throughput sequencing, and selected miRNAs related to LC or other tumors in the existing literature and databases to construct the circRNA/lncRNA-miRNA-mRNA network, and reveal the functions of ncRNAs. We speculated that we could study the functions and mechanisms of ncRNAs by means of well-known miRNAs from previous studies. For example, studies have reported that hsa-miR-4640-5p maybe the downstream target gene of hsa_circ_0044529, and may play important roles in the tumorigenesis of LC (45). Previous studies revealed that hsa-miR-608 could promote lung adenocarcinoma cell death and might be involved in the PI3K/AKT, Wnt, TGF-β, MAPK/ERK, and intrinsic pathways (46). From the data analysis, we can speculate that differentially expressed circRNAs and lncRNAs perform different functions by sponging multiple miRNAs. Furthermore, the ceRNA network may help us comprehend the mechanisms underlying the tumorigenesis and progression of LC.
In the GO and KEGG analyses, mRNA was selected as the main body of functional analysis, hoping to accurately predict the possible functions and signaling pathways of ncRNAs. In the GO analysis, the biological functions which were found to be most enriched were biological attachment, cell migration, cell adhesion, and cell proliferation, amongst others. These enrichment functions indicated that proliferation, invasion, and migration played important roles in the occurrence and development of LC. KEGG analysis revealed several signaling pathways that may be involved in the development of LC, including CAMs, the extracellular matrix receptor interaction pathway, and Adherens junction signaling pathways. GO and KEGG analyses results revealed the significance of cell proliferation and adhesion in the development of LC, which was consistent with the results of previous research (47). Through GO function enrichment and KEGG pathway analysis, the functions of differentially expressed ncRNAs and signaling pathways can be reliably predicted. These studies provide a scientific basis for the in-depth study of the pathogenesis of LC, and are conducive to the discovery of new tumor markers and the development of targeted treatment regimens.
In our study, SNHG29 was found to be significantly downregulated in LC cells and tissues and related to a better clinical outcome in LC patients. Specifically, considering that SNHG29 has been proved to inhibit the invasion and migration of laryngeal cancer through functional experiments, and that SNHG29 also inhibits the process of epithelial cell to mesenchymal transformation, we believe that SNHG29 can inhibit the metastasis of laryngeal cancer. In addition, through analysis of clinicopathological characteristics, we discovered that the downregulation of SNHG29 was correlated with differentiation level, T stage, lymphatic metastasis and clinical stage, indicating that SNHG29 may be considered an ideal biomarker for the prediction of prognosis. In the future, we can investigate novel medicine targeting the molecule of SNHG29 for precision treatment. And we can predict clinical outcome of patients with LC on the basis of understanding the SNHG29 expression level. In addition, increasing studies have revealed that dysregulated genes were correlated to risk stratification and local regional metastasis in tumors (M), combining with our results that SNHG29 was related to the clinicopathological characteristics and the metastasis of LC, we hypothesize that SNHG29 can be used as a potential biomarker for risk stratification and local regional metastasis in patients with LC, of course, this needs further verification. Our findings provide a better understanding of the molecular mechanisms of LC, which will facilitate the development of more effective diagnostic and therapeutic strategies.
Conclusions
This study lays the foundations for the discovery of many more transcripts using high-throughput sequencing of tissue samples. These transcripts are expected to become specific markers for the diagnosis and prognosis of LC. Further research on the functions of non-coding RNAs will contribute to the understanding of the pathogenesis of LC and provide a target for the accurate treatment of LC.
Acknowledgments
Funding: This work was supported by the Leading Talent of the “Hundred-Thousand-Ten Thousand Project” in Liaoning under grant no. XLYC1905004, and Shenyang High-level Innovation Talents Plan under grant no. RC190403.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-584
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-584
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-584). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All study participants provided informed consent, and the study was approved by the committee board of First Affiliated Hospital of China Medical University (number[2016]2016-30-2) The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Siegel RL, Miller KD. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Maasland DH, van den Brandt PA, Kremer B, et al. Alcohol consumption, cigarette smoking and the risk of subtypes of head-neck cancer: results from the Netherlands Cohort Study. BMC Cancer 2014;14:187. [Crossref] [PubMed]
- Niu J, Huang YJ, Wang LE, et al. Genetic polymorphisms in the PTPN13 gene and risk of squamous cell carcinoma of head and neck. Carcinogenesis 2009;30:2053-8. [Crossref] [PubMed]
- Bai Y, Hou J, Wang X, et al. Circ_0000218 plays a carcinogenic role in laryngeal cancer through regulating microRNA-139-3p/Smad3 axis. Pathol Res Pract 2020;216:153103 [Crossref] [PubMed]
- Zhang X, Wu N. LncRNA MEG3 inhibits cell proliferation and induces apoptosis in laryngeal cancer via miR-23a/APAF-1 axis. J Cell Mol Med 2019;23:6708-19. [Crossref] [PubMed]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature 2012;482:339-46. [Crossref] [PubMed]
- Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer 2017;16:94. [Crossref] [PubMed]
- Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol 2013;14:699-712. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Rybak-Wolf A, Stottmeister C, Glažar P, et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell 2015;58:870-85. [Crossref] [PubMed]
- Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci 2014;15:9331-42. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Xuan L, Qu L, Zhou H, et al. Circular RNA: a novel biomarker for progressive laryngeal cancer. Am J Transl Res 2016;8:932-9. [PubMed]
- Zhang G, Fan E, Zhong Q, et al. Identification and potential mechanisms of a 4-lncRNA signature that predicts prognosis in patients with laryngeal cancer. Hum Genomics 2019;13:36. [Crossref] [PubMed]
- Yue PJ, Sun YY, Li YH, et al. MYCT1 inhibits the EMT and migration of laryngeal cancer cells via the SP1/miR-629-3p/ESRP2 pathway. Cell Signal 2020;74:109709 [Crossref] [PubMed]
- Yang T, Li S, Liu J, et al. lncRNA-NKILA/NF-κB feedback loop modulates laryngeal cancer cell proliferation, invasion, and radioresistance. Cancer Med 2018;7:2048-63. [Crossref] [PubMed]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015;12:357-60. [Crossref] [PubMed]
- Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics 2009;25:1335-7. [Crossref] [PubMed]
- Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol 2015;16:4. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Yu H, Ye L, Mansel RE, et al. Clinical implications of the influence of Ehm2 on the aggressiveness of breast cancer cells through regulation of matrix metalloproteinase-9 expression. Mol Cancer Res 2010;8:1501-12. [Crossref] [PubMed]
- Fu C, Jiang L, Hao S, et al. Activation of the IL-4/STAT6 Signaling Pathway Promotes Lung Cancer Progression by Increasing M2 Myeloid Cells. Front Immunol 2019;10:2638. [Crossref] [PubMed]
- Ou R, Mo L, Tang H, et al. circRNA-AKT1 Sequesters miR-942-5p to Upregulate AKT1 and Promote Cervical Cancer Progression. Mol Ther Nucleic Acids 2020;20:308-22. [Crossref] [PubMed]
- Dacheng W, Songhe L, Weidong J, et al. LncRNA SNHG3 promotes the growth and metastasis of colorectal cancer by regulating miR-539/RUNX2 axis. Biomed Pharmacother 2020;125:110039 [Crossref] [PubMed]
- Ferbus D, Bovin C, Validire P, et al. The zinc finger protein OZF (ZNF146) is overexpressed in colorectal cancer. J Pathol 2003;200:177-82. [Crossref] [PubMed]
- Weigel C, Chaisaingmongkol J, Assenov Y, et al. DNA methylation at an enhancer of the three prime repair exonuclease 2 gene (TREX2) is linked to gene expression and survival in laryngeal cancer. Clin Epigenetics 2019;11:67. [Crossref] [PubMed]
- Li J, Sun S, Chen W, et al. Small Nucleolar RNA Host Gene 12 (SNHG12) Promotes Proliferation and Invasion of Laryngeal Cancer Cells via Sponging miR-129-5p and Potentiating WW Domain-Containing E3 Ubiquitin Protein Ligase 1 (WWP1) Expression. Med Sci Monit 2019;25:5552-60. [Crossref] [PubMed]
- Li G, Pan C, Sun J, et al. lncRNA SOX2-OT regulates laryngeal cancer cell proliferation, migration and invasion and induces apoptosis by suppressing miR-654. Exp Ther Med 2020;19:3316-24. [Crossref] [PubMed]
- Tsimplaki E, Argyri E, Sakellaridis A, et al. Oropharyngeal and laryngeal but not oral cancers are strongly associated with high-risk human papillomavirus in 172 Greek patients. J Med Virol 2017;89:170-6. [Crossref] [PubMed]
- Yang D, Shi Y, Tang Y, et al. Effect of HPV Infection on the Occurrence and Development of Laryngeal Cancer: A Review. J Cancer 2019;10:4455-62. [Crossref] [PubMed]
- Gong H, Shi Y, Zhou X, et al. Microbiota in the Throat and Risk Factors for Laryngeal Carcinoma. Appl Environ Microbiol 2014;80:7356-63. [Crossref] [PubMed]
- Papadimitrakopoulou V, Izzo JG, Liu DD, et al. Cyclin D1 and cancer development in laryngeal premalignancy patients. Cancer Prev Res (Phila) 2009;2:14-21. [Crossref] [PubMed]
- Gao M, Zhong A, Patel N, et al. High throughput RNA sequencing utility for diagnosis and prognosis in colon diseases. World J Gastroenterol 2017;23:2819-25. [Crossref] [PubMed]
- Arroyo M, Larrosa R. Expression-based, consistent biomarkers for prognosis and diagnosis in lung cancer. Clin Transl Oncol 2020;22:1867-74. [Crossref] [PubMed]
- Guan YF, Huang QL, Ai YL, et al. Nur77-activated lncRNA WFDC21P attenuates hepatocarcinogenesis via modulating glycolysis. Oncogene 2020;39:2408-23. [Crossref] [PubMed]
- Gong HL, Shi Y, Shi Y, et al. Reduced expression of mutS homolog 2 and mutL homolog 1 affects overall survival in laryngeal squamous cell carcinoma patients: Investigation into a potential cause. Oncol Rep 2013;30:1371-9. [Crossref] [PubMed]
- Zhou DD, Liu XF, Lu CW. Long non-coding RNA PVT1: Emerging biomarker in digestive system cancer. Cell Prolif 2017;50:e12398 [Crossref] [PubMed]
- Vo JN, Cieslik M, Zhang Y, et al. The Landscape of Circular RNA in Cancer. Cell 2019;176:869-81.e13. [Crossref] [PubMed]
- Lin SC, Lin CH. Cellular prion protein transcriptionally regulated by NFIL3 enhances lung cancer cell lamellipodium formation and migration through JNK signaling. Oncogene 2020;39:385-98. [Crossref] [PubMed]
- Liu G, Zhang Q, Xia L, et al. RNA-binding protein CELF6 is cell cycle regulated and controls cancer cell proliferation by stabilizing p21. Cell Death Dis 2019;10:688. [Crossref] [PubMed]
- Simonson B, Subramanya V, Chan MC, et al. DDiT4L promotes autophagy and inhibits pathological cardiac hypertrophy in response to stress. Sci Signal 2017;10:eaaf5967 [Crossref] [PubMed]
- Yan S, Zhou N, Zhang D, et al. PFKFB3 Inhibition Attenuates Oxaliplatin-Induced Autophagy and Enhances Its Cytotoxicity in Colon Cancer Cells. Int J Mol Sci 2019;20:5415. [Crossref] [PubMed]
- Yang R, Xing L, Zheng X, et al. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol Cancer 2019;18:4. [Crossref] [PubMed]
- Fan Y, Xia X, Zhu Y, et al. Circular RNA Expression Profile in Laryngeal Squamous Cell Carcinoma Revealed by Microarray. Cell Physiol Biochem 2018;50:342-52. [Crossref] [PubMed]
- Othman N, In LL, Harikrishna JA, et al. Bcl-xL silencing induces alterations in hsa-miR-608 expression and subsequent cell death in A549 and SK-LU1 human lung adenocarcinoma cells. PLoS One 2013;8:e81735 [Crossref] [PubMed]
- Shi C, Wu YY, Wei LQ. MiR-221 affects the proliferation and apoptosis of laryngeal cancer cells through the PI3K/AKT signaling pathway. Eur Rev Med Pharmacol Sci 2020;24:1258-63. [PubMed]


