
Homologous recombination repair gene mutations show no survival benefits in Chinese high-grade serous ovarian cancer patients
Introduction
Epithelial ovarian cancer is one of the most commonly diagnosed and lethal diseases among women, and 295,414 new cases and 184,799 deaths were documented in 2018 around the world (1). In China, 52,100 new cases and 22,500 deaths were reported in 2015 (2). Half of the patients will relapse within 16 months even after standard treatment including surgery and platinum-based chemotherapy. Furthermore, the 5-year overall survival (OS) rate is only approximately 50% (3,4).
BRCA1/2-associated ovarian cancers have been reported to have improved platinum sensitivity and OS (5,6). Several PARP inhibitors, such as Olaparib, Rucaparib, and Niraparib, have been approved by the Food and Drug Administration (FDA) as maintenance therapy for platinum-sensitive recurrence or as targeted therapy for BRCA-mutated patients. Somatic BRCA1/2 mutations are also included as the indication. Additionally, some reports show that patients with other homologous recombination repair (HRR) pathway gene mutations may also impact patients’ platinum sensitivity and prognosis, whereas other reports show inconsistent conclusions (7-9). Also, patients harbor some HRR genes may also respond to PARP inhibitor targeted therapy. However, data on the genetic burden of Chinese ovarian patients are limited. Thus, it is important to investigate the frequency of HRR gene mutations and their clinical-prognostic impact in Chinese ovarian cancer patients.
Since the vast majority of ovarian cancers are high-grade serous ovarian cancers (HGSC), we applied next-generation sequencing (NGS) in a consecutive Chinese HGSC patient cohort. Both germline and somatic deleterious mutations, covering BRCA1/2 and other 22 core HRR genes, were identified, and their relationships with clinical parameters and prognosis were analyzed. We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/atm-20-5136).
Methods
Clinical Data
This study was approved by the Committee at Fudan University Shanghai Cancer Center (IRB No. 1704171-6) and conducted in accordance with the Declaration of Helsinki (as revised in 2013). All individual participants signed the informed consent.
We enrolled 42 consecutive patients who underwent primary surgery for HGSC in November 2015 and December 2015 at our institution. Pathological data were primarily obtained from pathology reports. Then, all pathological slides were reviewed by two experienced gynecologic pathologists. HGSC were confirmed according to the WHO criteria.
Clinical data including age, FIGO stage, preoperative laboratory data, surgical outcomes, treatment strategy and patients’ disease status were obtained from medical records and cancer registries. Surgical outcomes were categorized as R0, R1 and R2 regarding the residual disease. Patients were clarified as platinum sensitive and platinum resistant, according to the response to platinum-based chemotherapy (10). Progression-free survival (PFS) was defined as the time interval from the date of primary surgery to the date of disease progression or recurrence, or the date of last follow-up with no relapsed disease. OS referred to the time interval from the date of the primary surgery to the date of death or the last follow-up (February, 2020).
HRR gene testing
Paired peripheral blood (or normal tissue) and tumor samples were taken at hospitals and sent to Burning Rock Laboratory, where DNA extraction, targeted DNA sequencing, variant calling, and interpretation were performed.
Sample collection and DNA extraction
2 mL peripheral blood collected in ethylenediaminetetraacetic acid (EDTA) tubes was centrifuged at 2,000 g for 10 min at 4 °C for intermediate white blood cell (WBC) layer collection. After resuspending in proteinase K/PBS solution, the genomic DNA of WBC was extracted by MagPure FFPE DNA LQ Kit (Magen, China). Tissue DNA was extracted using the QIAamp DNA FFPE tissue kit (Qiagen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The DNA concentration was measured using Qubit dsDNA assay (Thermo Fisher Scientific Inc. Waltham, MA, USA).
NGS library preparation
DNA shearing was performed using Covaris M220 followed by end repair, phosphorylation and adaptor ligation. Fragments of 200–400 bp in size were selected using Agencourt AMPure beads (Beckman Coulter, Brea, CA, USA) followed by hybridization with capture probe baits, hybrid selection with magnetic beads and polymerase chain reaction (PCR) amplification. A bioanalyzer high-sensitivity DNA assay was performed to assess the quality and size of the fragments. Fifty nanograms of DNA were used for library construction. Twelve PCR cycles were used for library amplification. Genetic profiles of all tumor samples were assessed by performing capture-based targeted deep sequencing using the HRD panel described below (Burning Rock Biotech Ltd., Guangzhou, China). The indexed samples were sequenced on Nextseq500 sequencer (Illumina, Inc., San Diego, CA, USA) with paired-end reads (read length 150 bp) and average sequencing depth of 680X. Paired WBC (n=41) or normal tissue (n=1) were used for germline mutation filtration.
Sequence data analysis
The sequence data were mapped to the human genome (hg19) using BWA aligner 0.7.10. Local alignment optimization, variant calling and annotation were performed using GATK 3.2, MuTect, and VarScan. Variants were filtered using the VarScan fpfilter pipeline, with loci with a depth less than 100 filtered out. At least 5 supporting reads were needed for INDELs; alternatively, 8 supporting reads were needed for SNVs. According to the ExAC, 1,000 Genomes, dbSNP, and ESP6500SI-V2 databases, variants with population frequency over 0.1% were grouped as SNP and excluded from further analysis. The remaining variants were annotated with ANNOVAR and SnpEff v3.6. A DNA translocation analysis was performed using both Tophat2 and Factera 1.4.3.
All gene variations detected were graded according to ACMG 2015 as follows: pathologic, likely pathologic, variation of uncertain significance, likely benign and benign. Deleterious HRR gene mutations included pathologic and likely pathologic mutations. It was defined as detailed functional defects or changes in gene expression.
Panel description
The HRD panel includes whole exons of 22 HRD-related genes (ATM, BARD1, BRIP1, CDK12, CHEK1, CHEK2, FANCI, FANCL, PPP2R2A, PALB2, RAD51C, RAD51D, RAD51B, RAD54L, ATR, EMSY, FANCA, FAM175A, NBN, MRE11A, RAD50, and PTEN) and whole exons and introns for BRCA1 and BRCA2. It also includes more than 10,000 SNPs from Agilent OneSeq for detecting loss of heterozygosity. The panel spans 2.8 MB of the human genome.
Statistical analyses
We used SPSS software (version 21.0) and GraphPad Prism (version 6.0) for the statistical analyses. Demographic data were described as the medians with ranges or the frequencies with percentages. Chi-squared or Fisher’s exact tests were used for comparison of the categorical data. The PFS and OS were analyzed with the Kaplan-Meier method with log-rank tests for univariate analyses, and cox regression analyses with hazard ratios (HRs) for the multivariate analyses. All P values reported were 2-sided, and P<0.05 was considered statistically significant, and.
Results
Patient characteristics and HRR gene mutations
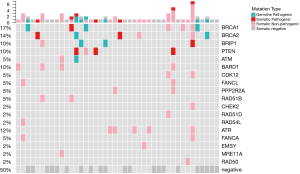
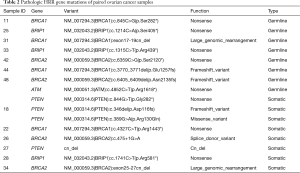
The patient characteristics are summarized in Table 1. The median (range) age of the patients was 58 [35–77]. The majority of the patients (41/42, 97.6%) had advanced stage (III–IV) HGSC. The HRR gene mutation profile is shown in Figure 1. Deleterious germline HRR-mutations were identified in 16.7% (7/42) of the HGSC patients. Among them, one patient had both germline BRCA2 and ATM mutations. Six patients had only somatic mutations, increasing the HRR mutation rates to 31.0% (13/42). Details of the gene mutations are listed in Table 2.

Full table
Full table
Treatment outcomes, survival analysis, and their correlations with HRR gene mutations
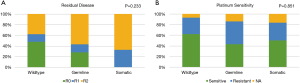
After primary staging or debulking surgery, 38.1% (16/42) of the patients had no residual disease. Thirty-eight out of 42 patients had adjuvant platinum-based chemotherapy. Among them, 24 (57.1%) were platinum sensitive (Table 1). In the HRR gene wild-type patients, 48.3% (14/29) of had no residual disease. However, the R0 rates in the germline (28.6%) and somatic (0%) HRR mutated groups were relatively lower (P=0.233, Figure 2A). The differences in platinum-sensitive rates among the three groups were not statistically significant (wild-type vs. germline vs. somatic, 62.1% vs. 42.9% vs. 50%, respectively, P=0.851, Figure 2B).
All patients were followed until February 2020, over four years. Five (11.9%) women experienced disease progression during adjuvant chemotherapy, and 22 (52.4%) patients exhibited documented recurrence. The median (95% CI) PFS was 22 (4.0–40.0) months, and 19 (45.2%) deaths were documented.
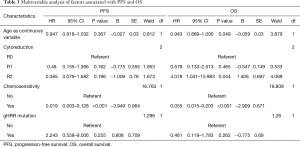
In the univariate analysis, germline HRR mutation was not associated with PFS (P=0.948, Figure 3A) or OS (P=0.321, Figure 3B). In the multivariate analysis with adjustments for age, surgical outcome and platinum sensitivity status, germline HRR mutation was not an independent predictor of PFS (HR =2.243, 95% CI, 0.558–9.006, P=0.255) or OS (HR =0.461, 95% CI, 0.119–1.783, P=0.262) (Table 3). In addition, no prognostic differences between somatic HRR mutated patients and wild-type patients were found.

Full table
Discussion
Our study showed that a considerable proportion of Chinese HGSC patients harbored HRR gene mutations. However, the HRR gene defect was not associated with improved survival in our cohort.
BRCA1/2 are critical HRR genes, and the reported prevalence varied among different ethnic populations (11-13). However, data on Chinese ovarian cancer patients are limited (14-16). Recently, studies investigating multiple HRR gene mutations have increased (8,17,18). Our study showed 31% deleterious HRR mutations, which is comparable with previous reports. This indicates a considerable proportion of patients with hereditary ovarian cancer and potential targeted therapy.
Previous reports have indicated that HRR gene mutations may impact platinum sensitivity and prognosis, but the conclusions were inconsistent (5-9). Pennington et al. (8) demonstrated that, except for germline BRCA1/2 mutations, somatic BRCA1/2 mutations and mutations of other homologous recombination genes have a similar positive impact on patients’ OS. Zhao et al. (9) showed that BRCA1/2-mutation carriers had favorable platinum sensitivity while mutations in other HR genes predicted a poor prognosis. In our study cohort, neither BRCA mutations nor HRR gene mutations were related to platinum sensitivity or prognosis. Shi’s study showed that BRCA mutation carriers had improved survival only in the subgroup with gross residual disease (19). In our study, patients with HRR mutations had a lower rate of complete or optimal cytoreduction, and this may also affect patients’ prognosis. Labidi-Galy (20) demonstrated that the location of mutation in the BRCA2 gene could affect the treatment efficacy and survival of patients, and only those with mutations located at RAD51-BD (exon 11) had prolonged PFS, platinum-free interval and OS. Thus, when assessing a patient’s individualized treatment, we should further explore the location and function of HRR mutations.
The main limitation of our study is the small sample size, and a slight change could influence the outcome observed. However, consecutive patients in two months were chosen to avoid selection bias. Notwithstanding this limitation, this study involved a group of homogeneous patients with the same histology and similar treatment strategies. Moreover, understanding HRR gene defects other than BRCA1/2 mutations could better elucidate the HGSC tumor gene profile.
Conclusions
Our results suggest that HRR gene defects are not associated with improved survival outcomes in our Chinese high-grade serous ovarian cancer patient cohort.
Acknowledgments
We would like to thank all doctors, nurses, patients, and their family members for their kindness to support our study.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/atm-20-5136
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5136). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Committee at Fudan University Shanghai Cancer Center (IRB No. 1704171-6) and conducted in accordance with the Declaration of Helsinki (as revised in 2013). All individual participants signed the informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Berek JS, Crum C, Friedlander M. Cancer of the ovary, fallopian tube, and peritoneum. International Journal Of Gynaecology And Obstetrics 2012;119 Suppl 2:S118-29. [Crossref] [PubMed]
- Feng Z, Wen H, Bi R, et al. A clinically applicable molecular classification for high-grade serous ovarian cancer based on hormone receptor expression. Sci Rep 2016;6:25408. [Crossref] [PubMed]
- Bolton KL, Chenevix-Trench G, Goh C, et al. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA 2012;307:382-90. [Crossref] [PubMed]
- Vencken PM, Kriege M, Hoogwerf D, et al. Chemosensitivity and outcome of BRCA1- and BRCA2-associated ovarian cancer patients after first-line chemotherapy compared with sporadic ovarian cancer patients. Ann Oncol 2011;22:1346-52. [Crossref] [PubMed]
- Ledermann JA, Drew Y, Kristeleit RS. Homologous recombination deficiency and ovarian cancer. Eur J Cancer 2016;60:49-58. [Crossref] [PubMed]
- Pennington KP, Walsh T, Harrell MI, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res 2014;20:764-75. [Crossref] [PubMed]
- Zhao Q, Yang J, Li L, et al. Germline and somatic mutations in homologous recombination genes among Chinese ovarian cancer patients detected using next-generation sequencing. J Gynecol Oncol 2017;28:e39. [Crossref] [PubMed]
- Feng Z, Wen H, Bi R, et al. Thrombocytosis and hyperfibrinogenemia are predictive factors of clinical outcomes in high-grade serous ovarian cancer patients. BMC Cancer 2016;16:43. [Crossref] [PubMed]
- Meindl A. Comprehensive analysis of 989 patients with breast or ovarian cancer provides BRCA1 and BRCA2 mutation profiles and frequencies for the German population. Int J Cancer 2002;97:472-80. [Crossref] [PubMed]
- Pal T, Permuth-Wey J, Betts JA, et al. BRCA1 and BRCA2 mutations account for a large proportion of ovarian carcinoma cases. Cancer 2005;104:2807-16. [Crossref] [PubMed]
- Sarantaus L, Vahteristo P, Bloom E, et al. BRCA1 and BRCA2 mutations among 233 unselected Finnish ovarian carcinoma patients. Eur J Hum Genet 2001;9:424-30. [Crossref] [PubMed]
- Li A, Xie R, Zhi Q, et al. BRCA germline mutations in an unselected nationwide cohort of Chinese patients with ovarian cancer and healthy controls. Gynecol Oncol 2018;151:145-52. [Crossref] [PubMed]
- Shi T, Wang P, Xie C, et al. BRCA1 and BRCA2 mutations in ovarian cancer patients from China: ethnic-related mutations in BRCA1 associated with an increased risk of ovarian cancer. Int J Cancer 2017;140:2051-9. [Crossref] [PubMed]
- Wu X, Wu L, Kong B, et al. The First Nationwide Multicenter Prevalence Study of Germline BRCA1 and BRCA2 Mutations in Chinese Ovarian Cancer Patients. Int J Gynecol Cancer 2017;27:1650-7. [Crossref] [PubMed]
- Eoh KJ, Kim JE, Park HS, et al. Detection of Germline Mutations in Patients with Epithelial Ovarian Cancer Using Multi-gene Panels: Beyond BRCA1/2. Cancer Res Treat 2018;50:917-25. [Crossref] [PubMed]
- Walsh T, Casadei S, Lee MK, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A 2011;108:18032-7. [Crossref] [PubMed]
- Shi T, Wang P, Tang W, et al. Survival Benefit of Germline BRCA Mutation is Associated with Residual Disease in Ovarian Cancer. Cell Physiol Biochem 2018;47:2088-96. [Crossref] [PubMed]
- Labidi-Galy SI, Olivier T, Rodrigues M, et al. Location of Mutation in BRCA2 Gene and Survival in Patients with Ovarian Cancer. Clin Cancer Res 2018;24:326-33. [Crossref] [PubMed]


