
Artificial intelligence for molecular neuroimaging
Introduction
Machine learning (ML) is a branch of AI that allows machines to perform tasks characteristically requiring human intelligence. Specifically, ML encompasses the set of algorithms and techniques enabling computers to process data without explicit programmed instructions dictating how the data is processed. Rather, the machine determines its own rules based on exposure to training data that can then be applied to process new data. For example, in terms of molecular brain imaging, ML algorithms might be used for applications such as: (I) generating computed tomography (CT) scans for attenuation correction (AC), (II) segmentation, (III) diagnosing disease or (IV) making outcome predictions.
ML algorithms are typically divided according to whether learning (i.e., training) is supervised, unsupervised or semi-supervised (reinforcement learning). In supervised learning an algorithm is given a training dataset that includes the classification or outcome for each item in the dataset so that the algorithm may infer distinguishing features. The idea is that once these distinguishing features are learned by the algorithm, the algorithm will be able to correctly classify new data according to the information learned from the input data. Common supervised algorithms are, for example, random forests (RFs) and support vector machines (SVMs). In unsupervised learning, the algorithm is provided with a dataset where the classification of the data is not given. The algorithm seeks common features and clusters the data according to these features. Common unsupervised algorithms include, for example, k-means clustering. In certain less common instances, algorithms may use a combination of supervised and unsupervised techniques. It is said that these algorithms have a semi-supervised learning approach.
Deep learning (DL) represents a subset of ML algorithms. In DL, the algorithms used are typically artificial neural networks (ANNs) where the ANNs are structured in such a way that input data is processed at several levels of abstraction. A greater number of levels translates into “deeper” (more advanced) learning. Convolutional neural networks (CNNs) are a type of ANN that is structured to process images, and algorithms that use CNNs are increasingly being applied to tasks in medical imaging. Further details regarding the principles of AI and ML are described elsewhere (1,2).
AI in molecular brain imaging is a rapidly evolving field with several notable publications across a broad spectrum of journals in the last few years (3-6). In the context of positron emission tomography (PET) and single photon emission computed tomography (SPECT), ML algorithms can be used to improve image quality, reduce scan time, delineate/segment brain tumors and metastases as well as assisting with image classification in the setting of neurodegenerative disease. This paper presents a review of recent applications of AI in molecular brain imaging, including neurodegenerative diseases, namely Alzheimer’s disease (AD) and Parkinson’s disease (PD), neuro-oncology, and other neurological abnormalities.
CT scan generation and AC
Today, attenuation and scatter correction for PET is typically performed using CT. In the absence of a concurrent CT, such as when dedicated brain PET scanners or scanners incorporating PET and magnetic resonance (MR) imaging (MRI) systems (PET/MR) are used, synthesis of CT data may be helpful for the purposes of attenuation and scatter correction.
A few studies have shown CT data can be derived from MR images using DL algorithms (7,8). In an exploratory study, a deep convolutional auto-encoder (CAE) network was trained to identify air, bone, and soft tissue using 30 MR co-registered to CT imaging and to generate synthetic CT data. The CAE is essentially a CNN with 13 convolutional layers that takes the MR as input, progressively reduces resolution, i.e., encode into features, followed by another 13 convolutional layers that progressively reconstruct images, i.e., decode, until a synthetic CT of original resolution is obtained. This algorithm was then evaluated in ten patients by comparing the synthesized CT to the acquired CT scans (8). The synthesized CT scans were then used to perform AC for an additional PET scans in five healthy subjects. The results were compared to standard MR- and CT-based AC. The authors found the PET reconstruction error was significantly lower using the DL algorithm than using conventional MR-based attenuation approaches (8). In another study, CT scans were generated by training a CNN model, also with encoding and decoding phases organized in a Unet-style structure (i.e., encoder layers followed by decoder layers), to map MR images to their corresponding CT images based upon CT and MR data from 18 patients with brain tumors (7). This method outperformed an atlas-based approach in less than one hundredth the amount of time.
CT data may also be derived from PET emission data. Arabi and Zaidi developed a method to predict AC factors using time-of-flight (TOF) PET emission data from [18F]FDG brain scans and DL (9). With their approach, TOF PET emission sinogram data was used to train a CNN to estimate AC factors with CT-based AC data serving as a reference. The estimated AC factors were then used for AC during PET image reconstruction and compared to a two-tissue class segmentation AC method (background-air and soft-tissue). This approach had significantly higher accuracy than the standard method, in terms of relative mean absolute error and root mean square error (9). This research group subsequently investigated assessing attenuation and scatter correction with only TOF PET emission for [18F]FDG, in comparison to [18F]DOPA, [18F]flortaucipir, and [18F]flutemetamol, which target dopamine receptors, tau pathology, and amyloid pathology, respectively (10). This work, based on the same CNN structure as in (9), showed moderate performance of their DL AC algorithm at generating PET attenuation corrected images, with <9% absolute standardized uptake value (SUV) bias for the radiotracers studied, and the best performance seen with [18F]FDG. Overall, this method was found to be vulnerable to outliers, still the most intriguing result shown is the greater bias observed with the radiotracers that have greater signal-to-noise ratios compared to [18F]FDG. Hwang et al. also used the recent advancement of TOF PET technology to augment their DL approach to AC when investigating patient datasets using the radiopharmaceutical 18F-fluorinated-N-3-fluoropropyl-2-β-carboxymethoxy-3-β-(4-iodophenyl)nortropane {[18F]FP-CIT} for brain dopamine transporter imaging (11). This radiopharmaceutical leads to high crosstalk artifacts and noise due to high uptake in the striatum leading to regions of high contrast against background. The aforementioned study aimed to overcome these limitations with DL. Based on TOF PET data alone, they employed three different CNN methods and found that a hybrid network combining aspects of CAE and Unet generated estimated linear coefficient attenuation maps similar to those determined by CT. Shiri et al. also aimed to obtain attenuation-corrected PET images directly from PET emission data and investigated [18F]FDG-PET brain images from 129 patients (12). Using a deep convolutional encoder-decoder architecture, their network was designed to map non-AC PET images to pixel-wise continuously valued measured AC PET images (12). Their approach achieved comparable results relative to CT-based AC images, and further demonstrated the potential of DL methods for emission-based AC of PET images.
Generative Adversarial Networks (GANs) are a recent DL innovation where two ANNs compete against each other, with the goal of learning how to generate images with characteristics similar to those in the training set. GANs have recently been used to generate AC images (13-15). Armanious et al. trained a GAN to generate pseudo CTs in a supervised approach that used pairs of [18F]FDG PET and their corresponding CT for training (13). The approach, which was found to have satisfactory accuracy, was tested by comparing PETs that were attenuation-corrected using both actual CTs and pseudo CTs.
AI in neuro-oncology
AI has been applied to molecular imaging in neuro-oncology to differentiate normal and abnormal images, provide automated tumor delineation and segmentation, predict survival outcomes, and aid in the development personalized treatment plans (16-24). The interpretation of [18F]FDG PET brain scans is challenging due to the high background glucose metabolism of gray matter which leads to low signal-to-noise ratios (25). AI has been shown to have the potential to mitigate this issue and improve the interpretation of [18F]FDG PET scans. CNNs use either two-dimensional (2D) or three-dimensional (3D) convolutional “kernels”, or matrixes, based on slice-to-slice data or a patch of all scans, respectively. 3D CNNs are computationally more intensive than 2D CNNs, yet are superior because they use voxel-wise rich data from 3D images which results in more accurate classification of scans. Nobashi et al. aimed to develop a 2D CNN based on a ResNet structure using multiple axes (transverse, coronal, and sagittal) with multiple window intensity levels to accurately classify [18F]FDG brain PET scans in cancer patients (16). This study showed that despite using less rich extraction of information from PET scans, 2D CNNs have the potential to differentiate normal and abnormal brain scans, particularly ensemble models (i.e., models that generate their overall classifications based on sub-classifications from multiple ML algorithms) with different window settings and axes compared to individual models.
Given that accurate tumor delineation and segmentation is critical in diagnosing and staging cancer, and that manual contouring is time-consuming and open to subjectivity, AI applications for automated tumor delineation is an attractive goal in oncology (17-21). Comelli et al. have done extensive work in recent years towards this goal, investigating various algorithms based on 2D PET slices to obtain the 3D shape of the tumor with [18F]FDG and carbon-11 labeled methionine {[11C]MET} radiotracers in brain PET images (26,27). Recently, the same research group proposed a PET reconstruction based on a 3D system in which the segmentation is performed by evolving an active surface directly into the 3D space (28). The relevance of this work in tumor delineation is that it performs shape reconstruction on the entire stack of slices simultaneously, naturally leveraging cross-slice data that could not be exploited in the 2D system. This fully 3D approach was evaluated in a data set of 50 [18F]FDG and [11C]MET PET scans of lung, head and neck, and brain tumors and a benefit was demonstrated with their novel approach, and though fully automated tumor segmentation was not achieved at this time, their future work incorporating ML components and CT data is intriguing. [11C]MET PET brain scans have also been the target of supervised ML to predict survival outcomes in glioma patients (22). In this study, 16 predictive ML models were investigated with combinations of in vivo features from [11C]MET PET scans, ex vivo features, and patient features for their ability to predict 36-month survival, with the most successful ML methods involving data from all three features with survival prediction sensitivities and specificities ranging from 86–98% and 92–95%, respectively (22).
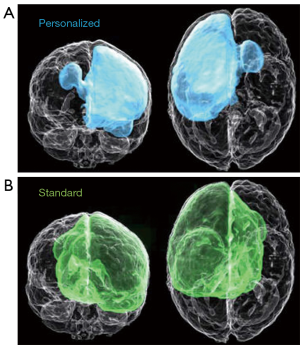
In the case of glioblastoma, treatment plans involve MRI to delineate tumors for resection followed by radiotherapy and chemotherapy to target residual and occult tumor cells. Unique to glioblastoma is the infiltration of occult cancer cells into the surrounding area that is not detected by MRI. Current treatment plans address this issue by uniformly extending the region of radiotherapy beyond the volume of the visible tumor, which does not address that the extent of occult infiltration is variable among glioblastoma patients (29,30). To address this issue, Lipkova et al. described a Bayesian ML framework that integrates data from MRI and PET scans with [18F]fluoroethyl-tyrosine {[18F]FET}, which has been shown to have uptake values proportional to tumor cell density and is much better at delineating tumor regions with occult cells than MRI (23). In a preliminary clinical research study, the radiotherapy plans generated from the inferred tumor cell infiltration maps obtained with the proposed Bayesian ML framework spare healthy tissue from unnecessary irradiation while yielding comparable accuracy with the standard radiotherapy protocol, as shown in Figure 1.
AI in neurodegenerative disease
AI has shown remarkable accuracy in diagnostic classification of neurodegenerative diseases such as AD and PD (4,6,31). Some approaches identify patterns and extract various features associated with AD and distinguish these from non-AD groups. In contrast, others aim to identify patterns in healthy controls and remove these from AD datasets. PET radiopharmaceuticals commonly used for studying AI applications in AD include [18F]FDG, to image glucose metabolism, as well as amyloid plaque imaging agents [18F]florbetapir, [18F]florbetaben, [11C]PiB, and [18F]flutemetamol, and tau imaging agents such as [18F]flortaucipir, among others. SPECT has also been studied for classifying AD with AI by examining cerebral blood flow, most commonly with the radiopharmaceutical 99m-technetium-ethyl cysteinate diethyl ester {[99mTc]ECD}.
Identifying patients with AD and mild cognitive impairment (MCI) with cerebral amyloid deposition may be helpful and has been studied using ML algorithms. This process is currently performed by a designated imaging analyst and is subject to human interpretation. Zukotynski et al. demonstrated the ability of RFs, a supervised ML algorithm, to successfully classify amyloid brain PET scans into positive or negative with sensitivity, specificity, and accuracy of 86%, 92%, and 90%, respectively (32). This approach used a 10,000-tree RF, where each tree was created using 15 randomly selected cases and 20 randomly selected features of SUV ratio per region of interest (ROI) (32). Zukotynski et al. next strived to identify ROIs in amyloid {[18F]florbetapir} and [18F]FDG PET scans that may be associated with a patient’s Montreal Cognitive Assessment (MoCA) score, again using RFs (33). In this study, RFs with 1,000 trees were constructed, with each tree using a random subset of 16 training [18F]florbetapir PET scans and 20 ROIs; the RFs identified ROIs associated with MoCA. Additionally, RFs with 1,000 trees were similarly constructed based on [18F]FDG PET scans, and identified specific ROIs associated with MoCA scores (33). Another study aiming to classify AD with the radiotracer [18F]florbetapir examined two structurally identical CNNs trained separately on MRI and amyloid PET data and compared the classification of each separately, as well as a different network that combined the two sets of data (34). This study determined that the greatest classification accuracy of 92%±2% was obtained with the CNN that used both MRI and amyloid PET data.
The majority of published studies focus on investigating a specific algorithm, hence comparing algorithms is challenging considering that the datasets in different studies vary in data acquisition, PET protocols, and data reconstructions. Brugnolo et al. addresses this gap in AI literature by reporting a head-to-head comparison of various automatic semi-quantification tools for their diagnostic performance based on brain [18F]FDG PET scans from prodromal AD patients and healthy controls (35). This aforementioned study compared three different statistical parametric mapping (SPM) approaches with a voxel-based tool, and a volumetric ROI SVM-based approach for their ability to differentiate prodromal AD from healthy control scans and found that the volumetric ROI-SVM-based approach outperformed all others studied with an area under the receiver operating characteristic (ROC) curve (AUC) of 0.95 compared to 0.84 and 0.83 for two different volumetric ROI-based methods, 0.79 for SPM maps, and 0.87 for the voxel-based tool (35).
In a slightly different approach to classifying AD, Choi et al. developed an unsupervised DL model, based on variational autoencoder, trained only by normal [18F]FDG brain PET scans, to define an “Abnormality Score” depending on how far a given patient scan was from the normal data (36). They measured the accuracy of their model using AUC analysis and report an AUC of 0.90 for differentiating AD from MCI data. The model was also validated on a heterogeneous cohort of patients with non-dementia related abnormalities, including abnormal behavioral symptoms and epilepsy, and found that experts’ visual interpretation was aided by the model in identifying non-dementia related abnormal patterns, in 60% of cases that did not initially identify as abnormal without the model (36).
AI has also been applied to molecular imaging data to predict patient outcomes. For example, to predict progression from MCI to dementia due to AD, Mathotaarachchi et al. implemented a random under sampling random forest (RUSRF) approach (37). This approach applied to the Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset provided a [18F]florbetapir prediction accuracy of 84% and an AUC of 0.91. A demonstration is available online at: http://predictalz.tnl-mcgill.com/PredictAlz_Amy.
As is common when data sets are limited in scope and size, many studies investigating the use of ML algorithms for automated prediction and classification of neurological disorders randomly divide their data into input “training” and “testing” groups; this is repeated many times for cross-validation, such that they are training their networks from the same pool of data they test with. This does not address whether or not the algorithm will be successful with new, completely unseen data from independently recruited participants (38). Improving the potential of ML algorithms to generalize complex neurological diseases outside their ‘training’ and ‘testing’ datasets can be done by incorporating more diverse data with ensemble models and may be beneficial when stratifying patients for therapeutic clinical trials. For example, amyloid beta deposition is a hallmark of AD, and is believed to start to develop many years before the onset of dementia (39). Several clinical trials attempting to target and lower Aβ levels in the brain in MCI and early dementia due to AD have failed. This failure may be partly attributed to patient selection. For example, in the negative bapineuzumab and solanezumab studies, a high percentage of patients with dementia but without increased cerebral Aβ levels were enrolled, and such patients may not be expected to benefit from the treatment (22). More recently, a positive amyloid PET scan is required for enrolment in anti-amyloid antibody trials. Often several data-types are included for the purpose of predicting outcome. Franzmeier et al. (38) applied a support vector regression to AD biomarkers from cerebrospinal fluid, structural MRI, amyloid-PET ([11C]PiB), and [18F]FDG data. Ensemble models combining multiple classifiers are known to improve prediction performance compared to individual classifiers (40). By incorporating data from 2 separate databases, Franzmeier et al. developed a robust algorithm that was able to predict the 4-year rate of cognitive decline in sporadic prodromal AD patients. The impact of such an algorithm is a substantial reduction in the sample size required in AD clinical trials, because proper stratification of patients who are most likely to benefit could be selected based on a positive amyloid PET scan. (38). Similarly, Ezzati et al. developed a ML model to predict Aβ status by including demographic, ApoE4 status, neuropsychological tests, MRI volumetric data and CSF biomarkers and training the algorithm with [18F]florbetapir brain PET scans from the ADNI database (41). The ML models that combined CSF biomarkers significantly outperformed neuropsychological tests and MR volumetrics alone. An ensemble model described by Wu et al. (42) combined three different classifiers using weighted and unweighted schemes improved the accuracy of predicting AD outcomes based on [11C]PiB brain PET images and determined that weighted ensemble models outperformed K-Nearest Neighbors, RFs, and ANNs. A novel, yet simple, approach of denoising using unsupervised ML was employed to improve signal-to-noise ratio of AD pattern expression scores from [18F]FDG brain PET scans, using 220 MCI patient scans available from ADNI (43). This work used principal component analysis—an unsupervised learning approach—to identify patterns of non-pathological variance in healthy control scans and removing them from the patient data before calculating the AD pattern expression score. Their pattern expression score-based prediction of AD conversion significantly improved from AUC 0.80 to 0.88. When combined with neuropsychological features and ApoE4 genotype data, the prediction model further improved with AUC 0.93 (43).
A few studies have examined combining [18F]FDG PET data with [99mTc]ECD SPECT data, which provides information about cerebral blood flow, to differentiate AD from healthy controls (44,45). Ferreira et al. compared applying a SVM model to data from [18F]FDG PET, [99mTc]ECD, and MRI images to determine which dataset would provide the greatest classification power (44). They achieved reasonable, though significantly greater accuracy with [18F]FDG PET data (68-71%), and [99mTc]ECD SPECT (68-74%), compared to MRI data (58%). In a later study, this research group examined applying a multiple kernel learning (MKL) method that would allow identification of the most relevant ROIs of classification based again on data from [18F]FDG PET, [99mTc]ECD SPECT, and MRI images (45). They compared using two different atlases to define ROIs from each imaging dataset, automated anatomical labeling and Brodmann’s area, and applied these to their MKL method, then compared these with the SVM model. They found much greater accuracy with their MKL method compared to SVM, the highest being for [18F]FDG PET with 92.50% accuracy found with both atlases (45).
AI has also been shown to have potential to classify PD using PET data with [18F]FDG or L-3,4-dihydroxy-6-[18F]fluorophenylalanine ([18F]DOPA), to assess degeneration in the dopaminergic pathway. Glaab et al. investigated the diagnostic power of two different ML methods, SVM and RF, on metabolic data with or without [18F]FDG or [18F]DOPA imaging, at differentiating PD from healthy controls (46). This study found that diagnostic power was lowest for any of these datasets alone, and that the greatest diagnostic power was obtained with combined [18F]DOPA and metabolic datasets (AUC =0.98). One limitation of AI includes overfitting of data, Shen et al. aimed to address this issue by adding the Group Lasso Sparse model to a Deep Belief Network (47). They applied their model to [18F]FDG PET scans from 2 cohorts of PD and healthy control subjects, the first cohort was randomly divided into training, validation, and test datasets, while the second cohort was used only to test the model. Their model outperformed traditional Deep Belief Networks at classifying [18F]FDG PET scans as PD or non-PD. Another study investigated extracting high-order radiomic features from [18F]FDG PET scans to differentiate PD from healthy controls (48). ROIs were identified based on the atlas-method; then features were selected by autocorrelation and the Fisher score algorithm. A SVM was trained on these extracted features to classify PD and healthy controls, which achieved 88.1%±5.3% accuracy compared to 80.1%±3.1% achieved with the traditional voxel values method (48).
SPECT is routinely performed to detect dopamine transporters with imaging agents such as 123-iodine-fluoropropylcarbomethoxyiodophenylnortropane ([123I]FP-CIT) (49). Several studies have investigated applying AI to [123I]FP-CIT SPECT datasets to achieve automated classification of PD. Choi et al. developed a CNN model trained on [123I]FP-CIT SPECT data from PD and healthy control subjects from the Parkinson’s Progression Markers Initiative (PPMI) database, then validated the model with an independent cohort of PD and non-parkinsonian tremor patients from the Seoul National University Hospital (50). Their model was able to overcome variability of human evaluation, and was able to accurately classify the atypical subgroup of PD in which dopaminergic deficit is absent in SPECT scans despite a clinical diagnosis of PD (50). Nicastro et al. also aimed to provide an automated classification model for PD combining semiquantitative striatal [123I]FP-CIT SPECT indices and SVM analysis in an approach that was able to differentiate PD from atypical and non-PD syndromes (51). In another study, researchers extracted seven features from the brain scans and assessed their ability to diagnose PD separately and in combination using SVM, k-nearest neighbors, and logistic regression (52). The greatest classification potential, 97.9% accuracy, 98% sensitivity, and 97.6% specificity, was found to be with SVM applied to all features simultaneously. In a similar study, [123I]FP-CIT SPECT scans were pre-processed using automated template-based registration followed by computation of the radiopharmaceutical binding potential at a voxel level, then using these data to classify images using a SVM model (53). This approach is currently being studied in clinical trials (ClinicalTrial.gov identifier NCT01141023).
Challenges, current status, and future prospects
The field of AI has grown substantially in recent years and has been applied to brain molecular imaging to improve the quality of images, reduce the amount of time a patient must be on a scanner, and assist in physician decision making and clinical interpretation of images. To improve the quality of images obtained from PET imaging systems that lack a concurrent CT scanner, AI has been applied to either MR or PET data to determine accurate AC and create synthetic CT data. By applying AI to PET data to obtain AC a patient may endure shorter scan times. AI has shown potential in assisting decision making in clinical care by automating the delineation of brain tumors and metastases from reconstructed imaging data, as well as automated classification of neurological diseases, such as AD and PD. Current clinical trials indicate that the use of AI in brain molecular imaging is indeed a growing field (ClinicalTrials.gov identifiers NCT00330109, NCT04357236, NCT04174287).
A rising trend in AI in brain molecular imaging is studies investigating ensemble models that involve multiple classifiers such as biomarkers and patient features. Neurological and neurodegenerative diseases are diagnosed by examining different features such as biomarkers in blood samples and scores on cognitive tests. Ensemble models that include these features with nuclear imaging data in their algorithms are showing greater classification and diagnostic potential compared to individual models. A gap exists in the literature regarding a lack of AI applied to novel radiopharmaceuticals; this is likely due to one of the major challenges of AI in nuclear imaging, which is the need for large training datasets. A typical clinical study with novel radiopharmaceuticals is unlikely to produce a large enough dataset to adequately train an AI algorithm. Another limitation is the potential biases of AI algorithms. Consider that the training, testing, and validation datasets are often selected randomly from the same larger dataset, this introduces bias into the validation of results. The ability of AI algorithms to perform tasks, such as the classification of images, and to have potential to be beneficial in a clinical setting is more evident if the models have been validated on independent cohorts from separate institutions. Additionally, it is difficult to compare AI algorithms used in different publications in the literature, for example, if considering algorithms for automated classification of AD, results will vary depending on the datasets used for validation. For this reason, the most robust studies on the development of new AI models will include a comparison to a standard AI model.
Despite these challenges, FDA-approved AI technologies are moving into clinics. Siemens has recently received FDA approval for use on their Biograph family of PET/CT imaging systems (54). These AI technologies, collectively named AIDAN, are comprised of propriety DL algorithms that allow four new features: FlowMotion AI, OncoFreeze AI, PET FAST Workflow AI, and Multiparametric Pet Suite AI. FlowMotion AI technology allows fast tailoring of PET/CT protocols in different patients by automatically detecting anatomical structures whose locations and sizes vary greatly between patients. PET FAST WorkFlow AI is applied to post-scan tasks to permit faster interpretation of results and reducing possibility of error. OncoFreeze AI applies AI to PET/CT acquisitions to correct for patient respiration motion, which can compromise image quality. Multiparametric PET Suite AI extracts arterial input function from PET/CT images to eliminate the pain and risk associated with arterial sampling during PET/CT scans. Together, these technologies permit faster, more accurate PET/CT acquisitions and image interpretations while allowing a more comfortable experience for the patient. Additionally, a distribution partnership was recently announced between CorTechs Labs and Subtle Medical involving Subtle Medical’s proprietary software, SubtleMRTM and SubtlePETTM, which are the first FDA-approved AI applications for medical imaging enhancement. These will be combined with CorTech Labs’ software, NeuroQuant® and PETQuantTM, which may allow PET/MRI facilities to reduce scan time without foregoing quantitative volumetric image analyses (55). SubtleMRTM improves image quality and denoises MR scans, while SubtlePETTM is able to denoise PET scans that are carried out in 25% of the time of a typical PET scan. NeuroQuant® and PETQuantTM are applications that provide automated and rapid analysis of MR and PET images, respectively. The reduced acquisition times with quantitative analysis of images will likely improve the patient experience and aid in data interpretation.
It is hard to say how the future of AI in molecular imaging will evolve. There remains the need for careful evaluation of the algorithms that are being used and developed, as well as where their use best fits in our clinical applications.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Dr. Steven P. Rowe) for the series “Artificial Intelligence in Molecular Imaging” published in Annals of Translational Medicine. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6220). The series “Artificial Intelligence in Molecular Imaging” was commissioned by the editorial office without any funding or sponsorship. Dr. SEB reports that she has received personal fees for 2 ad hoc consulting meetings for Hoffman LaRoche in 2018 and 2019. She has served on a speaker panel for an educational series on dementia diagnosis, imaging and management sponsored by Biogen in 2020. Her peer reviewed research on molecular imaging in dementia has received in kind support through provision of amyloid PET ligands from Lilly Avid, and GE Healthcare. Her institution has received funding for treatment trials. She conducts presymptomatic and prodromal Alzheimer Diseases from Eli-Lilly, Hoffman La Roche, Biogen and Eisai in the last 36 months. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Uribe CF, Mathotaarachchi S, Gaudet V, et al. Machine learning in nuclear medicine: part 1-introduction. J Nucl Med 2019;60:451-8. [Crossref] [PubMed]
- Tseng HH, Wei L, Cui S, et al. Machine learning and imaging informatics in oncology. Oncology 2020;98:344-62. [Crossref] [PubMed]
- Lohmann P, Kocher M, Ruge MI, et al. PET/MRI radiomics in patients with brain metastases. Front Neurol 2020;11:1. [Crossref] [PubMed]
- Liu X, Chen K, Wu T, et al. Use of multimodality imaging and artificial intelligence for diagnosis and prognosis of early stages of Alzheimer's disease. Transl Res 2018;194:56-67. [Crossref] [PubMed]
- Zaharchuk G. Next generation research applications for hybrid PET/MR and PET/CT imaging using deep learning. Eur J Nucl Med Mol Imaging 2019;46:2700-7. [Crossref] [PubMed]
- Duffy IR, Boyle AJ, Vasdev N. Improving PET imaging acquisition and analysis with machine learning: a narrative review with focus on Alzheimer's disease and oncology. Mol Imaging 2019;18:1536012119869070 [Crossref] [PubMed]
- Han X. MR-based synthetic CT generation using a deep convolutional neural network method. Med Phys 2017;44:1408-19. [Crossref] [PubMed]
- Liu F, Jang H, Kijowski R, et al. Deep learning MR imaging-based attenuation correction for PET/MR imaging. Radiology 2018;286:676-84. [Crossref] [PubMed]
- Arabi H, Zaidi H. Deep learning-guided estimation of attenuation correction factors from time-of-flight PET emission data. Med Image Anal 2020;64:101718 [Crossref] [PubMed]
- Arabi H, Bortolin K, Ginovart N, et al. Deep learning-guided joint attenuation and scatter correction in multitracer neuroimaging studies. Hum Brain Mapp 2020;41:3667-79. [Crossref] [PubMed]
- Hwang D, Kim KY, Kang SK, et al. Improving the accuracy of simultaneously reconstructed activity and attenuation maps using deep learning. J Nucl Med 2018;59:1624-9. [Crossref] [PubMed]
- Shiri I, Ghafarian P, Geramifar P, et al. Direct attenuation correction of brain PET images using only emission data via a deep convolutional encoder-decoder (Deep-DAC). Eur Radiol 2019;29:6867-79. [Crossref] [PubMed]
- Armanious K, Hepp T, Küstner T, et al. Independent attenuation correction of whole body [18F]FDG-PET using a deep learning approach with Generative Adversarial Networks. EJNMMI Res 2020;10:53. [Crossref] [PubMed]
- Hu Z, Li Y, Zou S, et al. Obtaining PET/CT images from non-attenuation corrected PET images in a single PET system using Wasserstein generative adversarial networks. Phys Med Biol 2020;65:215010 [Crossref] [PubMed]
- Dong X, Lei Y, Wang T, et al. Deep learning-based attenuation correction in the absence of structural information for whole-body positron emission tomography imaging. Phys Med Biol 2020;65:055011 [Crossref] [PubMed]
- Nobashi T, Zacharias C, Ellis JK, et al. Performance comparison of individual and ensemble CNN models for the classification of brain [18F]FDG-PET Scans. J Digit Imaging 2020;33:447-55. [Crossref] [PubMed]
- Berthon B, Marshall C, Evans M, et al. ATLAAS: an automatic decision tree-based learning algorithm for advanced image segmentation in positron emission tomography. Phys Med Biol 2016;61:4855-69. [Crossref] [PubMed]
- Berthon B, Evans M, Marshall C, et al. Head and neck target delineation using a novel PET automatic segmentation algorithm. Radiother Oncol 2017;122:242-7. [Crossref] [PubMed]
- Huang B, Chen Z, Wu PM, et al. Fully Automated delineation of gross tumor volume for head and neck cancer on PET-CT using deep learning: a dual-center study. Contrast Media Mol Imaging 2018;2018:8923028 [Crossref] [PubMed]
- Blanc-Durand P, Van Der Gucht A, Schaefer N, et al. Automatic lesion detection and segmentation of [18F]FET PET in gliomas: a full 3D U-Net convolutional neural network study. PLoS One 2018;13:e0195798 [Crossref] [PubMed]
- Blanc-Durand P, Van Der Gucht A, Verger A, et al. Voxel-based [18F]FET PET segmentation and automatic clustering of tumor voxels: a significant association with IDH1 mutation status and survival in patients with gliomas. PLoS One 2018;13:e0199379 [Crossref] [PubMed]
- Papp L, Pötsch N, Grahovac M, et al. Glioma survival prediction with combined analysis of in vivo [11C]MET PET features, ex vivo features, and patient features by supervised machine learning. J Nucl Med 2018;59:892-9. [Crossref] [PubMed]
- Lipkova J, Angelikopoulos P, Wu S, et al. Personalized radiotherapy design for glioblastoma: integrating mathematical tumor models, multimodal scans, and bayesian inference. IEEE Trans Med Imaging 2019;38:1875-84. [Crossref] [PubMed]
- Kebir S, Weber M, Lazaridis L, et al. Hybrid [11C]MET PET/MRI combined with "machine learning" in glioma diagnosis according to the revised glioma WHO classification 2016. Clin Nucl Med 2019;44:214-20. [Crossref] [PubMed]
- Wong TZ, van der Westhuizen GJ, Coleman RE. Positron emission tomography imaging of brain tumors. Neuroimaging Clin N Am 2002;12:615-26. [Crossref] [PubMed]
- Comelli A, Stefano A, Russo G, et al. A smart and operator independent system to delineate tumours in positron emission tomography scans. Comput Biol Med 2018;102:1-15. [Crossref] [PubMed]
- Comelli A, Stefano A, Bignardi S, et al. Active contour algorithm with discriminant analysis for delineating tumors in positron emission tomography. Artif Intell Med 2019;94:67-78. [Crossref] [PubMed]
- Comelli A, Bignardi S, Stefano A, et al. Development of a new fully three-dimensional methodology for tumours delineation in functional images. Comput Biol Med 2020;120:103701 [Crossref] [PubMed]
- Stupp R, Brada M, van den Bent MJ, et al. High-grade glioma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol 2014;25:iii93-101. [Crossref] [PubMed]
- Burnet NG, Thomas SJ, Burton KE, et al. Defining the tumour and target volumes for radiotherapy. Cancer Imaging 2004;4:153-61. [Crossref] [PubMed]
- Garali I, Adel M, Bourennane S, et al. Histogram-based features selection and volume of interest ranking for brain PET image classification. IEEE J Transl Eng Health Med 2018;6:2100212 [Crossref] [PubMed]
- Zukotynski K, Gaudet V, Kuo PH, et al. The use of random forests to classify amyloid brain PET. Clin Nucl Med 2019;44:784-8. [Crossref] [PubMed]
- Zukotynski K, Gaudet V, Kuo PH, et al. The use of random forests to identify brain regions on amyloid and [18F]FDG-PET associated with MoCA score. Clin Nucl Med 2020;45:427-33. [Crossref] [PubMed]
- Punjabi A, Martersteck A, Wang Y, et al. Neuroimaging modality fusion in Alzheimer's classification using convolutional neural networks. PLoS One 2019;14:e0225759 [Crossref] [PubMed]
- Brugnolo A, De Carli F, Pagani M, et al. Head-to-head comparison among semi-quantification tools of brain [18F]FDG-PET to aid the diagnosis of prodromal Alzheimer's disease. J Alzheimers Dis 2019;68:383-94. [Crossref] [PubMed]
- Choi H, Ha S, Kang H, et al. Deep learning only by normal brain PET identify unheralded brain anomalies. EBioMedicine 2019;43:447-53. [Crossref] [PubMed]
- Mathotaarachchi S, Pascoal TA, Shin M, et al. Identifying incipient dementia individuals using machine learning and amyloid imaging. Neurobiol Aging 2017;59:80-90. [Crossref] [PubMed]
- Franzmeier N, Koutsouleris N, Benzinger T, et al. Predicting sporadic Alzheimer's disease progression via inherited Alzheimer's disease-informed machine-learning. Alzheimers Dement 2020;16:501-11. [Crossref] [PubMed]
- Jack CR Jr, Knopman DS, Jagust WJ, et al. Tracking pathophysiological processes in Alzheimer's disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol 2013;12:207-16. [Crossref] [PubMed]
- Rokach L. Ensemble-based classifiers. Artif Intell Rev 2010;33:1-39. [Crossref]
- Ezzati A, Harvey DJ, Habeck C, et al. Predicting amyloid-β levels in amnestic mild cognitive impairment using machine learning techniques. J Alzheimers Dis 2020;73:1211-9. [Crossref] [PubMed]
- Wu W, Venugopalan J, Wang MD. 11C-PIB PET image analysis for Alzheimer's diagnosis using weighted voting ensembles. Annu Int Conf IEEE Eng Med Biol Soc 2017;2017:3914-7. [Crossref] [PubMed]
- Blum D, Liepelt-Scarfone I, Berg D, et al. Controls-based denoising, a new approach for medical image analysis, improves prediction of conversion to Alzheimer's disease with [18F]FDG-PET. Eur J Nucl Med Mol Imaging 2019;46:2370-9. [Crossref] [PubMed]
- Ferreira LK, Rondina JM, Kubo R, et al. Support vector machine-based classification of neuroimages in Alzheimer's disease: direct comparison of [18F]FDG-PET, rCBF-SPECT and MRI data acquired from the same individuals. Braz J Psychiatry 2017;40:181-91. [Crossref] [PubMed]
- Rondina JM, Ferreira LK, de Souza Duran FL, et al. Selecting the most relevant brain regions to discriminate Alzheimer's disease patients from healthy controls using multiple kernel learning: a comparison across functional and structural imaging modalities and atlases. Neuroimage Clin 2017;17:628-41. [Crossref] [PubMed]
- Glaab E, Trezzi JP, Greuel A, et al. Integrative analysis of blood metabolomics and PET brain neuroimaging data for Parkinson's disease. Neurobiol Dis 2019;124:555-62. [Crossref] [PubMed]
- Shen T, Jiang J, Lin W, et al. Use of overlapping group LASSO sparse deep belief network to discriminate Parkinson's disease and normal control. Front Neurosci 2019;13:396. [Crossref] [PubMed]
- Wu Y, Jiang JH, Chen L, et al. Use of radiomic features and support vector machine to distinguish Parkinson's disease cases from normal controls. Ann Transl Med 2019;7:773. [Crossref] [PubMed]
- Hustad E, Aasly JO. Clinical and imaging markers of prodromal Parkinson's disease. Front Neurol 2020;11:395. [Crossref] [PubMed]
- Choi H, Ha S, Im HJ, et al. Refining diagnosis of Parkinson's disease with deep learning-based interpretation of dopamine transporter imaging. Neuroimage Clin 2017;16:586-94. [Crossref] [PubMed]
- Nicastro N, Wegrzyk J, Preti MG, et al. Classification of degenerative parkinsonism subtypes by support-vector-machine analysis and striatal [123I]FP-CIT indices. J Neurol 2019;266:1771-81. [Crossref] [PubMed]
- Oliveira FPM, Faria DB, Costa DC, et al. Extraction, selection and comparison of features for an effective automated computer-aided diagnosis of Parkinson's disease based on [123I]FP-CIT SPECT images. Eur J Nucl Med Mol Imaging 2018;45:1052-62. [Crossref] [PubMed]
- Oliveira FP, Castelo-Branco M. Computer-aided diagnosis of Parkinson's disease based on [123I]FP-CIT SPECT binding potential images, using the voxels-as-features approach and support vector machines. J Neural Eng 2015;12:026008 [Crossref] [PubMed]
- FDA clears AIDAN artificial intelligence for Siemens Healthineers Biograph PET/CT Portfolio [press release]. Siemens Healthineers, 2020.
- CorTechs Labs and Subtle Medical announce distribution partnership [press release]. San Diego: Business Wire, 2020.


