
Analysis of microRNA expression in cerebral ischemia/reperfusion after mild therapeutic hypothermia treatment in rats
Introduction
Cerebral ischemia has always been the main cause of mortality and disability globally. Only tissue plasminogen activator delivered within a limited time window and mild hypothermia are considered to be effective neuroprotectants (1-3). Mild hypothermia treatment has been reported to reduce neuronal apoptosis by suppressing the disfunction of mitochondria and blood brain barrier, proliferation of microglia cells and production of superoxide anions and nitric oxide (4-6). A significant neuroprotection was obtained when hypothermia was started within therapeutic time window of 4 h after reperfusion in rat model (6). Furthermore, in patients with middle cerebral artery infarction, therapeutic hypothermia treatment can significantly reduce intracranial pressure, alleviate cerebral edema, and improve neurological dysfunction (7,8). Furthermore, a number of studies have investigated the potential for combination of mild hypothermia with drug therapy, which was proved to be more effective in protecting neurons than hypothermia alone in an experimental model of cerebral ischemia (6).
However, the molecular and cellular processes involved in the treatment of mild hypothermia, especially posttranscriptional regulation, have yet to be illuminated.
MicroRNAs (miRNAs), small noncoding single-stranded RNA molecules of 18–23 nucleotides long, are important to negatively regulate the gene expression. After integrating with the 3'-untranslated region of messenger RNA (mRNA), they inhibit the translation or promote the degradation of mRNA (9,10). MiRNAs have been reported to play critical roles in the development, homoeostasis, and pathological processes of the central nervous system (11,12). Numerous studies of middle cerebral artery occlusion (MCAO) models have demonstrated that abnormal miRNA expression is involved in the pathogenic progress and treatment of cerebral ischemia. Many miRNAs have been acknowledged to be differentially expressed in the circulating blood of stroke patients, and regarded as diagnostic and prognostic markers (13-16).
Although the expression of miRNAs have been reported to be involved in angiogenesis, apoptosis, and regeneration after cerebral ischemia in previous studies. However, miRNA profiles in mild hypothermia treatment after cerebral ischemia still remained indeterminate (17). This study aimed at describing the miRNA expression profiles related to mild hypothermia treatment after cerebral ischemia. Thus, we anticipate to get a new perspective in the exploration of effective treatments for cerebral ischemia. For instance, a correlation has been verified between inhibition of miRNA-15b (miR-15b) and the protective effects of mild therapeutic hypothermia in MCAO mice (18).
In this study, the miRNA expression profiles of MCAO rats treated with normothermia and mild hypothermia were analyzed. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed and the functions of the target genes were elucidated. Additionally, pathway analysis tools were applied to analyze the signaling pathways of the theses genes. Then, an analysis of potential miRNA targets in mild hypothermia treatment of MCAO was carried out to explore the detailed mechanism and provide a novel target for mild hypothermia therapy in cerebral ischemia.
We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-21-143).
Methods
Animal experiments
The experimental procedures were approved by the Institutional Animal Care and Use Committee of Capital Medical University (Beijing, China) and conducted in accordance with the principles outlined in the National Institutes of Health (US) Guide for the Care and Use of Laboratory Animals.
Anesthesia was induced in Male Sprague-Dawley rats (weight: 280–310 g) through inhalation of 5% isoflurane in oxygen. The rats were then subjected to tracheal intubation and mechanical ventilation with 2% isoflurane in 30% O2/70% N2. Focal cerebral ischemia was induced by MCAO as previously described (19). The right common artery and external carotid artery were exposed, and the right middle artery was occluded through the insertion of 3-0 monofilament nylon suture into the internal carotid artery. After MCAO, the cerebral blood flow was monitored by color flow Doppler. The rats were randomly assigned into a sham-operated group and a MCAO group (n=4–5/per group). For miRNA microarray analysis, the sham-operated and MCAO groups were treated with mild hypothermia or normothermia. Blood pressure was monitored using a data acquisition system (MP100A-CE, BIOPAC Systems, Inc., Santa Barbara, CA, USA). The body temperature of the rats was monitored using a thermocouple probe inserted into the rectum and was maintained at 37.0±0.5 °C during the operation with a temperature-regulated heating and a heating lamp. The rats’ cerebral temperature was measured using a thermocouple probe placed under the right temporalis. Mild hypothermia (33±0.5 °C) was induced from the onset of ischemia and maintained for 3 hours through the placement of ice packs around the head/neck, which were replaced every hour.
Intraventricular injection
MiRNA mimics and inhibitor (n=7–8 per group) for intracerebroventricular injection were purchased from Life Technologies. The injection was performed 60 minutes prior to cerebral ischemia as previously described (20). A needle was used to stereotaxically implant 10 μM of miRNA in a total volume of 10 μL mixture, including 2 μL Lipofectamine 2000, into the left ventricle (−0.8 mm anterior, +1.5 mm lateral, and −3.2 mm ventral relative to the bregma).
Infarct volume analysis
Infarct volume was calculated and analyzed by 2,3,5-triphenyltetrazolium chloride (TTC) as previously described (21). Six coronal brain sections were sliced to a thickness of 2 mm using a brain matrix. The brain sections were incubated in 2% TTC-saline solution for 30 minutes at 37 °C, then fixed in 4% paraformaldehyde/phosphate-buffered saline. The sections were photographed 24 hours after fixation and analyzed with Image-Pro Plus Analysis Software (Media Cybernetics Corporation, USA).
Measurement of neuronal apoptosis via terminal deoxynucleotidyl transferase dUTP nick end labeling
A terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) test kit (Qiming Biotech Co., Ltd., Shanghai, China, OX02752) was utilized to measure neuronal apoptosis. Brain tissues were fixed on glass slides with 4% paraformaldehyde for 20 minutes and permeabilized with 0.1% (v/v) Triton™ X-100 for 5 minutes. Then, the slides were incubated with TUNEL incubation mixture in a dark and humid box at 37 °C for 1.5 hours and stained with 4’,6-diamidino-2-phenylindole (DAPI). The TUNEL apoptotic cells were observed and counted under a fluorescence microscope.
miRNA microarray
miRNA microarray was proceeded with the 7th generation of miRCURYTM LNA Array (v.18.0) (Exiqon) which contains 3100 capture probes, covering the miRNAs information in miRbase of human, mouse, and rats, as well as all viral miRNAs related to these species. Additionally, 25 miRPlus™ human miRNAs are also included in this arrat.
miRNA extraction
Total RNA was extracted from rat brain tissues with TRIzol (Invitrogen) and miRNeasy mini kit (Qiagen, Hilden, Germany) as described in manufacturer’s instructions. A Nanodrop spectrophotometer (ND-1000, Nanodrop Technologies, Wilmington, Delaware) was used to assess the RNA concentration and quality, and the integrity of the RNA was measured using denaturing agarose gel electrophoresis.
miRNA labeling and array hybridization
After the measurement of RNA quality and quantity, the miRCURY™ Hy3™/Hy5™ Power labeling kit (Exiqon, Vdebaek, Denmark) was used to label RNA according to the manufacturer’s guideline. Briefly, 1.0 μg total RNA and 2 µL water were combined with 1.0 µL calf-intestinal alkaline phosphatase (CIP) (Exiqon, Vdebaek, Denmark) and CIP buffer. After incubation for 30 minutes at 37 °C and 5 minutes at 95 °C, 3.0 µL labeling buffer, 1.5 µL fluorescent label (Hy3TM), 2.0 µL dimethyl sulfoxide (DMSO), and 2.0 µL labeling enzyme were mixed and incubated for 60 minutes at 16 °C, and 15 minutes at 65 °C to terminate the reaction.
Next, the hybridization of Hy3-labeled samples to the miRCURY™ LNA Array (v.18.0) was proceeded. 5 µL Hy3-labeled samples and 25 µL hybridization buffer were mixed and denatured for 2 minutes at 95 °C, then incubated on ice for 2 minutes, and hybridized to microarray for 16–20 hours at 56 °C in 12-Bay Hybridization System (Hybridization System; Nimblegen Systems, Inc., Madison, WI, USA). After several washes with a wash buffer kit (Exiqon), The slides were washed with wash buffer kit (Exiqon) and dried by centrifugation for 5 minutes at 400 rpm. The slides were scanned with the Axon GenePix 4000B microarray scanner (Axon Instruments, Foster City, CA, USA).
Grid alignment and data analysis of the scanned images were performed by GenePix Pro 6.0 software (Axon). The normalization factor were calculated by miRNAs samples with intensity ≥30. After normalization, significantly differentially expressed miRNAs were identified through volcano plot filtering. Upregulated or downregulated miRNAs were defined with those with fold change ≥1.5 and P≤0.05. Hierarchical clustering analysis was performed using MEV software (v4.6, TIGR).
Quantitative reverse transcription-polymerase chain reaction (RT-qPCR)
Total RNA was extracted from rat brain tissue using TRIzol (Qiagen, Hilden, Germany) according to the manual. Then, the quantity of total RNA was determined by 1.5% formaldehyde denaturing agarose gel electrophoresis. RNA was reverse-transcribed to cDNA with a stem-loop RT primer using M-MLV reverse transcriptase (Promega, Madison, WI, USA). Quantitative PCR was carried out using AmpliTap Gold Enzyme (Applied Biosystems, Austin, TX, USA). RT reactions were performed for 10 minutes at 16 °C, 30 minutes at 37 °C, and 5 minutes at 65 °C. Real-time PCR amplification was performed as follows: 95 °C for 10 minutes, 40 cycles at 95 °C for 15 seconds, 60 °C for 30 seconds, and 70 °C for 30 seconds.
Functional and pathway enrichment analyses
The functions of genes including biological process (BP), cellular component (CC), and molecular function (MF), were investigated by GO (http://www.geneontology.org) analysis. Fisher’s exact test was used to find if there was more overlap between the differentially expressed gene list and the GO annotation list than would be expected by chance. The significance of GO term enrichment among the differentially expressed genes was denoted by the P value: the lower the P value, the more significant the GO term (P value ≤0.05 is recommended).
The signal pathway enrichment of target genes was examined by KEGG (http://www.genome.ad.jp/kegg/) pathways analysis. The significance of the related pathway was denoted by the P value: the lower the P value, the more significant the pathway (a P value cut-off of 0.05 is recommended). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
miRNA-mRNA network
The associations between all differentially expressed miRNAs (DEmiRNAs) and their targets were predicted through data integration using miRbase (www.mirbase.org/), miRDB (http://mirdb.org), and miRanda (http://www.microrna.org/microrna/home.do). Subsequently, the network map was established by a Cytoscape software (version 2.8.3; http://cytoscape.github.io/).
Statistical analysis
Data were expressed as means ± standard deviation. The statistical analysis of difference of infarct volume and neuronal apoptosis between the mimic-treated group and the inhibitor-treated group was performed using independent sample t tests by GraphPad Prism 8.0 (GraphPad Software Inc., La Jolla, CA, USA). P values <0.05 was considered statistically significant.
Results
Total RNA quality
Total RNA was extracted from the ipsilateral cortex of MCAO rats treated with normothermia, at 4 and 24 hours after the induction of mild hypothermia respectively. The integrity of the RNA revealed sharp 28S and 18S bands in a ratio greater than or close to 1:1. RNA purity and concentration were measured using a NanoDrop spectrophotometer, and an A260/A280 ratio above 1.8 was recommended for good array performance.
Identification of DEmiRNAs
At 4 hours after mild therapeutic hypothermia, 6 upregulated miRNA and 6 downregulated miRNAs were identified between hypothermic and normothermic rats. At 24 hours after mild therapeutic hypothermia, 41 upregulated miRNA and 10 downregulated miRNAs were identified between the hypothermia and normothermia groups. A volcano plot and hierarchical analysis of the DEmiRNAs are presented in Figure 1.

RT-qPCR confirmation of the DEmiRNAs
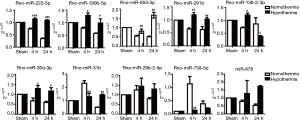
RT-qPCR was performed to confirm the miRNA-seq results. The 9 most significantly upregulated miRNAs at 24 hours after mild therapeutic hypothermia were selected, including rno-miR-291b, rno-miR-878, rno-miR-653-3p, rno-miR-222-5p, rno-miR-29b-2-5p, rno-miR-31b, rno-miR-30d-3p, rno-miR-190b-5p, and rno-miR-138-2-3p. One downregulated miRNA, rno-miR-758-5p, was also selected. The results of RT-qPCR were consistent with those of miRNA-seq (Figure 2).
GO and KEGG analysis of DEmiRNAs
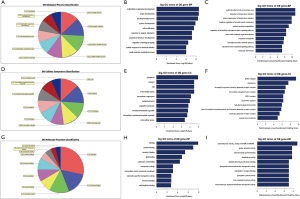
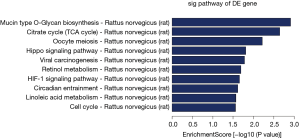
The biological functions of the selected miRNAs were analysed using GO and KEGG pathway analysis. The 10 most significantly enriched GO terms, determined according to their P values, are displayed in Figure 3. The most significantly enriched terms for BP, CC, and MF were multicellular organism development (P=2.7964E-09), cytoplasm (P=1.14045E-05), and binding (P=1.3743E-08), respectively. Furthermore, the DEmiRNAs were found to be involved in 16 subclasses of KEGG pathways, with the mucin type O-glycan biosynthesis pathway being the most significantly enriched pathway (Figure 4).
miRNA-mRNA network
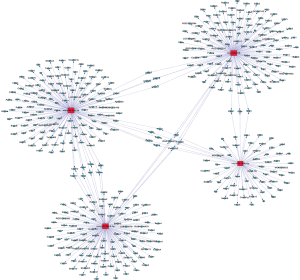
The target mRNAs of DEmiRNAs were analyzed using miRBase, miRanda, and miRDB. The miRNA-mRNA network was constructed based on target pairs. In the network, the top 4 miRNAs regulating the highest number of target genes were miR-29b-2-5p, miR-190b-5p, miR-878, and miR-30d-3p (Figure 5).
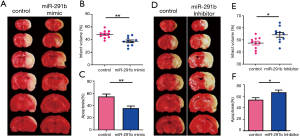
miR291b overexpression reduced the infarct volume and neuronal apoptosis in MCAO under normothermic conditions
From the DEmiRNAs, miR-291b was selected, and its effects in MCAO rats were investigated. An injection of miR-291b mimic or inhibitor was administered to the ipsilateral ventricle of rats 60 minutes before MCAO surgery under normothermia. Infarct volume and neuronal apoptosis were compared 24 hours after reperfusion. As shown in Figure 6, miR-291b overexpression significantly decreased the infarct volume and neuronal apoptosis in rats treated with miR-291b mimic compared with the control group (P<0.05) (Figure 6A,B,C). In contrast, miR-291b inhibition significantly increased the infarct volume and neuronal apoptosis in rats with cerebral ischemia treated with miR-291b inhibitor (Figure 6D,E).
Discussion
Cerebral ischemia/reperfusion injuries trigger a complex pathological process, which includes a number of gene expression changes. Recently, miRNAs were proven to be important regulators in neuronal apoptosis following cerebral ischemia/reperfusion, and further study revealed the roles of miRNAs in mild hypothermia treatment. The present study aimed to use microarray analysis to detect the miRNA profiles of rats with cerebral ischemia after treatment with mild hypothermia. At 4 hours after mild hypothermia, 6 miRNAs were upregulated and 6 were downregulated in the ipsilateral ischemic cortex, and after 24 hours, 41 and 10 miRNAs were upregulated and downregulated, respectively. Then, DEmiRNAs were subjected to GO and KEGG analysis. Our results revealed that the DEmiRNAs were mainly involved in the regulation of multicellular organism development and that the mucin type O-glycan biosynthesis pathway was the most enriched pathway. Then, the miRNA-target network was constructed, and the degrees of miR-29b-2-5p, miR-190b-5p, miR-878, and miR-30d-3p were revealed to be higher than those of other miRNAs.
In recent studies, miR-29 family members have been identified as critical regulators in brain development, cerebral ischemia, and neurodegenerative diseases (22-24). The expression of miR-29b has been demonstrated to show significant upregulation, not only in the brains of MCAO rats, but also in neurons and astrocytes exposed to oxygen and glucose deprivation (OGD). Furthermore, miR-29b was revealed to up-regulate the expression of myeloid cell leukemia-1 (MCL-1) and B-cell lymphoma 2 (BCL-2) proteins following down-regulation of caspase-3 expression in a cerebral I/R injury cell model, which was confirmed to result from the regulation of the Akt signaling pathway by miR-29b (25-28). Additionally, it has been confirmed that repression of BCL-2 or Bcl2L2 gene occurred after the upregulation of miR-29b, which subsequently resulted in the promotion of neuronal cell death (27,29). Interestingly, miR-29b has been reported to be significantly downregulated in the blood of stroke patients, with its downregulation potentially being a predictor of poor prognosis through associations with infarct volume aggravation, edema, and blood-brain barrier disruption via the regulation of aquaporin-4 (30), 12-lipoxygenase (31), and inflammatory response after cerebral ischemia (32). However, the role and mechanism of miR-29b in cerebral ischemia need further investigations.
The miR-29 family was also found to be downregulated in Alzheimer's disease (AD) patients, resulting in increased expression of β-Site APP cleavage enzyme 1 (BACE1), amyloid plaque formation, and neuronal cell apoptosis (33,34). Moreover, miR-29 family members have also been reported to bear a correlation with synapse formation and synaptic plasticity through their targeting of actin-related protein 2/3 complex subunit 3 (ARPC3), and axon guidance through their regulation of neuron navigator 3 (NAC3) (35,36). Han et al.’s study showed that the expression of miR-29b in PD patients with dementia was significantly downregulated compared with that in patients with mild cognitive impairment. Moreover, the downregulation of miR-29b was significantly higher in PD patients with mild cognitive impairment compared with those with no cognitive impairment. Therefore, miR-29b downregulation is associated with cognitive impairment. It is possible that the miR-29 family can regulate neuronal survival, communication, and homeostasis, which play vital roles in cognitive impairment. The damage of mir-29 to PD cognition may be mediated by synaptic regulator PARK7 (DJ-1), mitogen activated protein 6 and 7, memory conversion regulator (CREB), nerve growth and neurotrophic factors, which need to be further verified (37).
miR-190, located on the proximal end of the long arm of 15 (15q22.2), has 2 main mature forms: miR-190-5p and miR-190-3p. The role of miR-190 in the regulation of genes and pathways related to the tumorigenesis and progression of various cancers has been shown. Recent studies discovered the significantly different expression and roles of miR-190 in traumatic brain injury and neurodegenerative diseases (38,39). Pei et al. subjected the mouse hippocampal neuronal cell line HT-22 to OGD to mimic ischemia injury and found that miR-190b expression in astrocytes was significantly higher than that in astrocyte exosomes. Astrocyte exosome-mediated transfer attenuated the inflammation and neuronal apoptosis induced by OGD by suppressing autophagy. Furthermore, the neurotropic gene Atg7 was identified as the target gene and confirmed to regulate the expression of interleukin (IL)-1β, IL-6, and tumor necrosis factor-alpha (TNF-α) (40). Sun et al. reported downregulation of miR-190 in a 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced Parkinson’s Disease (PD) mouse model and in lipopolysaccharide (LPS)-induced BV2 cells. However, upregulation induced by miR-190 mimics inhibited the inflammatory response, with reduced expression of inducible Nitric-Oxide Synthase (iNOS), IL-6, TNF-α, and transforming growth factor beta 1 (TGF-β1), and increased expression of anti-inflammatory mediators, such as IL-10, in BV2 cells. NOD-like receptor family, pyrin domain containing 3 (NLRP3) as subsequently identified as the target gene of miR-190, and the inflammatory response and neuronal apoptosis were alleviated through negative regulation of Nlrp3 in the MPTP-induced PD mouse model (41).
miR-30d-5p has been found to be downregulated in brains with hypoxia-ischemia (HI) and in serum following cerebral ischemia. Increased expression of miR-30d-5p has been reported to result in reduced autophagy and increased neuronal apoptosis. Moreover, inhibition by antagomir was found to decrease the infarct volume and improve neurological function after HI injury (42,43). In rat cortical astrocytes subjected to OGD to mimic HI conditions, autophagy activity was observed to be enhanced while cell apoptosis was significantly decreased by miR-30d inhibition. However, the effects observed with inhibition of miR-30d were reversed by knockdown of Beclin-1, suggesting that the autophagy and apoptosis regulated by miR-30d in the in OGD-treated astrocytes was mediated by targeting Beclin-1 (44).
In addition to Beclin1, miR-30d has also been found to regulate the expression of Atg5. In primary microglial cells, miR-30d-5p was observed to promote the suppression of autophagy and alleviate the OGD induced inflammatory response by targeting Beclin-1 and Atg5. Furthermore, exosomes from miR-30d-5p enriched adipose-derived stem cells significantly reduced the inflammation infarction area following MCAO, which resulted from autophagy suppression mediated by the promotion of microglial polarization toward the M1 phenotype (43).
Mucins are a class of highly O-glycosylated glycoproteinsfound on the surface of or secreted by cells. Mucin-type O-glycans are formed by O-glycans with α linked to serine or threonine residues in mucins, which is initiated with N-acetylgalactosamine. Altered glycosylation has been established as a hallmark in many cancers. Mucin-type O-glycan biosynthesis and altered mucin-type O-glycans serve crucial roles in determining the tumor phenotype and adhesive properties, as well as in malignant invasion and metastasis (45,46). However, the role of mucin-type O-glycans in diseases of the nervous system have yet to be clarified.
miR-291b-3p is a member of the miR-290 cluster, which has been demonstrated the important miRNAs in murine embryonic stem cells (ESCs) (47,48). miR-291b-3p has been explored to mediate the metabolism of lipids and glucose in liver through targeting p65 to regulate phosphatase and tensin homolog deleted on chromosome ten (PTEN) expression and adenosine monophosphate-activated kinase α1 (AMPKα) (49,50). Additionally, miR-291b-3p was demonstrated to promote hepatocyte apoptosis via downregulating the expression of RNA-binding protein Hu antigen R (HuR). Moreover, HuR was also identified as target gene of miR-291b-3p involved in the endothelial cell apoptosis and dysfunction with increased expression of Vascular cell adhesion molecule-1 (VCAM-1) in EOMA cells treated with H2O2 (51,52). In our study, intraventricular injection of miR-291b significantly reduced the infarct volume and neuronal apoptosis, however the neuroprotective effect was revered by the injection of miR-291b inhibitor. MiR-291b was presumably meditated in the neuronal apoptosis in MACO rat after reperfusion. Further studies on the molecular mechanisms by which miR291b alleviates the neuronal apoptosis should be carried out.
The ischemia-reperfusion cascade involved during the pathological of cerebral ischemia is a complex series of events, which could be triggered by multiple cellular pathways that act coordinately, and miRNAs simultaneously regulate numerous target genes (53). However, the mechanisms that result changes of miRNA transcription after cerebral ischemia are not known. The expressions of the RNases Drosha and Dicer, the Drosha cofactor Pasha, and the pre-miRNA transporter exportin-5 demonstrated no significant changes in cerebral ischemia, which are essential to regulate the biogenesis of miRNA (54).
Conclusions
In summary, we observed significant miRNA expression changes in rats after the induction of mild therapeutic hypothermia following MCAO. The biological functions, downstream signaling pathways, and associated target genes of involved miRNAs were systematically analyzed for the first time. Our findings provide a preliminary clue for investigating the pathological hallmarks of mild hypothermia treatment for cerebral ischemia at the miRNA-mRNA level. Upregulation of miR-291b exhibited neuroprotective effects in MCAO rats after reperfusion and is likely to be a promising novel therapeutic strategy. However, our findings need to be verified in further investigations.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (Grant no. 81471209, 81871021) and the National Natural Science Foundation for Distinguished Young Scholars (Grant no. 81325007).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-143
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-143
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-143). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All animal experiments were approved by the Institutional Animal Care and Use Committee of Capital Medical University (Beijing, China) (No. 201802002) and conducted in accordance with the principles outlined in the National Institutes of Health (US) Guide for the Care and Use of Laboratory Animals. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mozaffarian D, Benjamin EJ, Go AS, et al. Heart disease and stroke statistics-2015 update: a report from the American Heart Association. Circulation 2015;131:e29-e322. [Crossref] [PubMed]
- Muresanu DF, Buzoianu A, Florian SI, et al. Towards a roadmap in brain protection and recovery. J Cell Mol Med 2012;16:2861-71. [Crossref] [PubMed]
- Kurisu K, Yenari MA. Therapeutic hypothermia for ischemic stroke; pathophysiology and future promise. Neuropharmacology 2018;134:302-9. [Crossref] [PubMed]
- Choi HA, Badjatia N, Mayer SA. Hypothermia for acute brain injury—mechanisms and practical aspects. Nat Rev Neurol 2012;8:214-22. [Crossref] [PubMed]
- Jadhav AP, Jovin TG. Endovascular therapy for acute ischemic stroke: the standard of care. Brain Circ 2016;2:178-82. [Crossref] [PubMed]
- Han Z, Liu X, Luo Y, et al. Therapeutic hypothermia for stroke: Where to go? Exp Neurol 2015;272:67-77. [Crossref] [PubMed]
- Cattaneo G, Meckel S. Review of selective brain hypothermia in acute ischemic stroke therapy using an intracarotid, closed-loop cooling catheter. Brain Circ 2019;5:211-7. [Crossref] [PubMed]
- Natarajan G, Laptook A, Shankaran S. Therapeutic hypothermia: How can we optimize this therapy to further improve outcomes? Clin Perinatol 2018;45:241-55. [Crossref] [PubMed]
- Nelson P, Kiriakidou M, Sharma A, et al. The microRNA world: small is might. Trends Biochem Sci 2003;28:534-40. [Crossref] [PubMed]
- Chen PY, Meister G. microRNA-guided posttranscriptional gene regulation. Biol Chem 2005;386:1205-18. [Crossref] [PubMed]
- Meza-Sosa KF, Pedraza-Alva G, Pérez-Martínez L. microRNAs: key triggers of neuronal cell fate. Front Cell Neurosci 2014;8:175. [Crossref] [PubMed]
- Lopez-Ramirez MA, Reijerkerk A, de Vries HE, et al. Regulation of brain endothelial barrier function by microRNAs in health and neuroinflammation. FASEB J 2016;30:2662-72. [Crossref] [PubMed]
- Bao J, Zhou S, Pan S, et al. Molecular mechanism exploration of ischemic stroke by integrating mRNA and miRNA expression profiles. Clin Lab 2018;64:559-68. [Crossref] [PubMed]
- Tiedt S, Prestel M, Malik R, et al. Seq identifies circulating miR125a-5p, miR-125b-5p and miR-143-3p as potential biomarkers for acute ischemic stroke. Circ Res 2017;121:970-80. [Crossref] [PubMed]
- Dewdney B, Trollope A, Moxon J, et al. Circulation microRNAs as biomarkers for acute ischemic stroke: a systemic review. J Stroke Cerebrovasc Dis 2018;27:522-30. [Crossref] [PubMed]
- Martinez B, Peplow PV. Blood microRNAs as potential diagnostic and prognostic markers in cerebral ischemic injury. Neural Regen Res 2016;11:137-58. [Crossref] [PubMed]
- Tan JR, Koo YX, Kaur P, et al. MicroRNAs in stroke pathogenesis. Curr Mol Med 2011;11:76-92. [Crossref] [PubMed]
- Liu X, Wen S, Zhao S, et al. Mild therapeutic hypothermia protects against cerebral ischemia/reperfusion injury by inhibiting miR-15b expression in rats. Aging Dis 2018;9:401-11. [Crossref] [PubMed]
- Belayev L, Alonso OF, Busto R, et al. Middle cerebral artery in the rat by intraluminal suture. Neurological and pathological evaluation of an improved model. Stroke 1996;27:1616-22. [Crossref] [PubMed]
- McCarthy CA, Vinh A, Callaway JK, et al. Angiotensin AT2 receptor stimulation causes neuroprotection in a conscious rat model of stroke. Stroke 2009;40:1482-9. [Crossref] [PubMed]
- Suzuki Y, Chen F, Marchal G, et al. Microplasmin reduces ischemic brain damage and improves neurological function in a rat stroke model monitored with MRI. Stroke 2004;35:2402-6. [Crossref] [PubMed]
- Roshan R, Shridhar S, Sarangdhar MA, et al. Brain-specific knockdown of miR-29 results in neuronal cell death and ataxia in mice. RNA 2014;20:1287-97. [Crossref] [PubMed]
- Yang G, Song Y, Zhou X, et al. MicroRNA-29c targets β-site amyloid precursor protein-cleaving enzyme 1 and has a neuroprotective role in vitro and in vivo. Mol Med Rep 2015;12:3081-8. [Crossref] [PubMed]
- Hébert SS, Horré K, Nicolaï L, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc Nat Acad Sci 2008;105:6415-20. [Crossref] [PubMed]
- Ziu M, Fletcher L, Rana S, et al. Temporal differences in microRNA expression patterns in astrocytes and neurons after ischemic injury. PLoS One 2011;6:e14724. [Crossref] [PubMed]
- Di Y, Lei Y, Yu F, et al. MicroRNAs expression and function in cerebral ischemia reperfusion injury. J Mol Neurosci 2014;53:242-50. [Crossref] [PubMed]
- Rong W, Yang L, Li CY, et al. MiR-29 inhibits neuronal apoptosis in rats with cerebral infarction through regulating Akt signaling pathway. Eur Rev Med Pharmacol Sci 2020;24:843-50. [PubMed]
- Huang Z, Lu L, Jiang T, et al. miR-29b affects neurocyte apoptosis by targeting MCL-1 during cerebral ischemia/reperfusion injury. Exp Ther Med 2018;16:3399-404. [Crossref] [PubMed]
- Shi G, Liu Y, Liu T, et al. Upregulated miR-29b promotes neuronal cell death by inhibiting Bcl2L2 after ischemic brain injury. Exp Brain Res 2012;216:225-30. [Crossref] [PubMed]
- Zhang YZ, Wang J, Xu F, et al. Circulating miR-29b and miR-424 as prognostic markers in patients with acute cerebral infarction. Clin Lab 2017;63:1667-74. [Crossref] [PubMed]
- Wang Y, Huang J, Ma Y, et al. MicroRNA-29b is a therapeutic target in cerebral ischemia associated with aquaporin 4. J Cereb Blood Flow Metab 2015;35:1977-84. [Crossref] [PubMed]
- Khanna S, Rink C, Ghoorkhanian R, et al. Loss of miR-29b following acute ischemic stroke contributes to neural cell death and infarct size. J Cereb Blood Flow Metab 2013;33:1197-206. [Crossref] [PubMed]
- Zong Y, Wang H, Dong W, et al. miR-29c regulates BACE1 protein expression. Brain Res 2011;1395:108-15. [Crossref] [PubMed]
- Kole AJ, Swahari V, Hammond S, et al. miR-29b is activated during neuronal maturation and targets BH3-only genes to restrict apoptosis. Genes Dev 2011;25:125-30. [Crossref] [PubMed]
- Zong Y, Yu P, Cheng H, et al. miR-29c regulates NAV3 protein expression in a transgenic mouse model of Alzheimer’s disease. Brain Res 2015;1624:95-102. [Crossref] [PubMed]
- Lippi G, Steinert JR, Marczylo EL, et al. Targeting of the Arpc3 actin nucleation factor by miR-29a/b regulates dendritic spine morphology. J Cell Biol 2011;194:889-904. [Crossref] [PubMed]
- Han L, Tang Y, Bai X, et al. Association of the serum micro-29 family with cognitive impairment in Parkinson’s disease. Aging (Albany NY) 2020;12:13518-28. [Crossref] [PubMed]
- Yu Y, Cao XC. miR-190-5p in human disease. Cancer Cell Int 2019;19:257. [Crossref] [PubMed]
- Meissner L, Gallozzi M, Balbi M, et al. Temporal profile of MicroRNA expression in contused cortex after traumatic brain injury in mice. J Neurotrauma 2016;33:713-20. [Crossref] [PubMed]
- Pei X, Li Y, Zhu L, et al. Astrocyte-derived exosomes transfer miR-190b to inhibit oxygen and glucose deprivation-induced autophagy and neuronal apoptosis. Cell Cycle 2020;19:906-17. [Crossref] [PubMed]
- Sun Q, Wang S, Chen J, et al. MicroRNA-190 alleviates neuronal damage and inhibits neuroinflammation via Nlrp3 in MPTP-induced Parkinson’s disease mouse model. J Cell Physiol 2019;234:23379-87. [Crossref] [PubMed]
- Zhao F, Qu Y, Zhu J, et al. miR-30d-5p plays an important role in autophagy and apoptosis in developing rat brains after hypoxic-ischemic injury. J Neuropathol Exp Neurol 2017;76:709-19. [Crossref] [PubMed]
- Jiang M, Wang H, Jin M, et al. Exomes from miR-30d-5p-ADSCSs reverse acute ischemic stroke-induced, autophagy-mediated brain injury by promoting M2 microglial/macrophage polarization. Cell Physiol Biochem 2018;47:864-78. [Crossref] [PubMed]
- Zhao F, Qu Y, Wang H, et al. The effect of miR-30d on apoptosis an autophagy in cultured astrocytes under oxygen-glucose deprivation. Brain Res 2017;1671:67-76. [Crossref] [PubMed]
- Staudacher E. Mucin-Type O-Glycosylation in invertebrates. Molecules 2015;20:10622-40. [Crossref] [PubMed]
- Kudelka MR, Ju T, Heimburg-Molinaro J, et al. Simple sugars to complex disease--mucin-type O-glycans in cancer. Adv Cancer Res 2015;126:53-135. [Crossref] [PubMed]
- Kaspi H, Chapnik E, Levy M, et al. Brief report: miR-290-295 regulate embryonic stem cell differentiation propensities by repressing Pax6. Stem cells 2013;31:2266-72. [Crossref] [PubMed]
- Lichner Z, Páll E, Kerekes A, et al. The miR-290-295 cluster promotes pluripotency maintenance by regulating cell cycle phase distribution in mouse embryonic stem cells. Differentiation 2011;81:11-24. [Crossref] [PubMed]
- Guo J, Dou L, Meng X, et al. Hepatic MiR-291b-3p Mediated Glucose Metabolism by Directly Targeting p65 to Upregulate PTEN Expression. Sci Rep 2017;7:39899. [Crossref] [PubMed]
- Meng X, Guo J, Fang W, et al. Liver MicroRNA-291b-3p Promotes Hepatic Lipogenesis through Negative Regulation of Adenosine 5'-Monophosphate (AMP)-activated Protein Kinase α1. J Biol Chem 2016;291:10625-34. [Crossref] [PubMed]
- Sui X, Yu S, Dou L, et al. miR-291b-3p mediated ROS-induced endothelial cell dysfunction by targeting HUR. Int J Mol Med 2018;42:2383-92. [Crossref] [PubMed]
- Guo J, Li M, Meng X, et al. MiR-291b-3p induces apoptosis in liver cell line NCTC1469 by reducing the level of RNA-binding protein HuR. Cell Physiol Biochem 2014;33:810-22. [Crossref] [PubMed]
- Hu Y, Deng H, Xu S, et al. MicroRNAs regulate mitochondrial function in cerebral ischemia-reperfusion injury. Int J Mol Sci 2015;16:24895-917. [Crossref] [PubMed]
- Dharap A, Bowen K, Place R, et al. Transient focal ischemia induces extensive temporal changes in rat cerebral microRNAome. J Cereb Blood Flow Metab 2009;29:675-87. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)


