
Molecular profiling of Chinese systemic anaplastic large cell lymphoma patients: novel evidence of genetic heterogeneity
Introduction
Anaplastic large cell lymphoma (ALCL) refers to a group of CD30-positive T-cell non-Hodgkin lymphomas (1,2). ALCL, which accounts for 13.8% of all peripheral T-cell lymphomas (PTCLs) (3), can be classified as primary cutaneous ALCL (pcALCL) and systemic ALCL. According to the World Health Organization (WHO) classification of Tumors of Hematopoietic and Lymphoid Tissues, systemic ALCL can be further divided based on anaplastic lymphoma kinase(ALK) status as ALK-negative ALCL and ALK-positive ALCL, the identification of which depends on either immunohistochemistry (IHC) or fluorescence in situ hybridization (FISH) (1).
In recent years, gene rearrangement or overexpression of ALK has been considered to be a favorable prognostic biomarker for ALCL patients (4,5). Approximately half of the patients diagnosed with ALCL have ALK-positive status and have high response to standard chemotherapy regimen consisting of cyclophosphamide, doxorubicin, vincristine, and prednisone or prednisolone (CHOP) (6). Contrastingly, due to inefficacy of standard regimens, patients with ALK-negative ALCL often undergo more intensive therapies than ALK-positive ALCL patients (7-9). A growing number of studies have reported that the 5-year survival of a subset of ALK-negative ALCL patients with dual specificity phosphatase 22 (DUSP22) rearrangement was comparable to that of ALK-positive ALCL patients who received the same treatment regimen (9-11). Routine stem cell transplantation may lead to overtreatment for subsets of ALK-negative ALCL patients, such as those with DUSP22 rearrangement, resulting in unnecessary increases in costs and risks for those patients. Rearrangements involving the tumor protein p63 (TP63) is considered to be a biomarker of poor prognosis for ALK-negative ALCL patients, with a 5-year survival rate of only 17% (10).
The National Comprehensive Cancer Network (NCCN) Clinical Practice Guidelines in Oncology for T-Cell lymphomas recommend that molecular analysis is used to detect DUSP22 rearrangement if ALK-negative ALCL is diagnosed and follow the same recommended treatment regimen as that for ALK-positive patients (10-13). Illumination of the genetic and clinical heterogeneity of ALCL patients may facilitate improvements in the clinical management of ALCL patients. However, due to the rarity of ALCL, the genetic heterogeneity of this disease has not been well investigated. In this study, we retrospectively enrolled 36 systemic ALCL patients and performed genetic profiling of their tumor tissue samples using capture-based targeted next-generation sequencing (NGS) with a panel spanning 112 lymphoma-related genes, in order to elucidate the somatic profiles of ALCL patients with distinct clinical features. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7574).
Methods
Patient and study design
This retrospective study included 36 patients who were diagnosed with systemic ALCL according to the WHO classification of Tumors of Hematopoietic and Lymphoid Tissue (2008) between January, 2015 and July, 2018 in Fujian Medical University Cancer Hospital and Fujian Cancer Hospital. All procedures involving human participants were performed in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol was approved by the Ethics Committee of Fujian Medical University Cancer Hospital and Fujian Cancer Hospital (approval number: 2017-055-01). Written informed consent was provided by all the patients included in the study.
Patient clinical data including age, sex, anatomic site, sites of extra nodal involvement, skin involvement during the disease course, disease stage, first-line treatment, treatment response, survival, and immunophenotype [CD3 antigen (CD3), Ki67 antigen (Ki67), BCL2 apoptosis regulator (BCL2), BCL6 transcription repressor (BCL6), or tumor necrosis factor (TNF) receptor superfamily member 8 (CD30)] were collected. Treatment response was assessed by the investigators according to the response evaluation criteria in lymphoma (RECIL) of the International Working Group (14). ALK status was detected by performing IHC. The threshold for immunopositivity was ≥20% of tumor cells (CD30 ≥80%). FISH was performed to detect DUSP22 rearrangement. Capture-based targeted NGS with a panel spanning 112 lymphoma-related genes (Burning Rock Biotech, Cat. No. LK205) was performed on tissue samples collected at baseline. Overall survival (OS) was evaluated from the time of initial diagnosis to death, or last follow-up in surviving patients.
DNA extraction and library preparation
DNA was extracted from formalin-fixed paraffin-embedded (FFPE) tissue samples using a QIAamp DNA FFPE tissue kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The DNA concentration was determined using the Qubit dsDNA assay (Life Technologies, Carlsbad, CA, USA). The M220 Focused-ultrasonicator (Covaris, Woburn, MA, USA) was used for DNA fragmentation, which was followed by end repair, phosphorylation, and adaptor ligation. DNA fragments of 200 to 400 bp were selected with AMPure beads (Agencourt AMPure XP kit; Beckman Coulter, Brea, CA, USA), and hybridization with capture probe baits, hybrid selection with magnetic beads, and polymerase chain reaction amplification were subsequently performed. A high-sensitivity DNA assay was used to examine the quality of the DNA (Bioanalyzer 2100; Agilent Technologies, Santa Clara, CA, USA). Indexed samples were sequenced using a Nextseq500 sequencer (Illumina, Inc., Hayward, CA, USA) with paired-end reads.
Sequencing data analysis
Sequencing data were mapped to the human genome (hg19) using Burrows-Wheeler aligner version 0.7.10 (15). Local alignment optimization, variant calling, and annotation were performed with GATK version 3.2 (Broad Institute, Cambridge, MA, USA) (16), and VarScan version 2.4.3 (Genome Institute, Washington University, Washington, DC, USA) (17). Loci with a depth of less than 100 were filtered out using the VarScan filter pipeline. At least five and eight supporting reads were required for insertions or deletions (INDELs) and single-number variations (SNVs), respectively. For all targeted regions, the aimed average sequencing depth was 2,000×. Variants with a population frequency >0.1% in public databases, including Exome Aggregation Consortium, 1,000 Genomes Project, ESP6500SI-V2 and dbSNP, were categorized as single-nucleotide polymorphisms (SNPs) and were excluded from further analyses. The ANNOVAR (18) and SnpEff version 3.6 (Wayne State University, Detroit, MI, USA) (19) softwares was used to annotate the remaining variants. Factera version 1.4.3 was employed for DNA translocation analysis (20).
Copy number variation (CNV) analysis was performed based on the depth of coverage data of capture intervals and corrected against sequencing bias resulting from GC content and probe design. The coverage of different samples was normalized to comparable scales based on the average coverage of all captured regions. The limits for CNV detection were 1.5 and 2.64 for deletions and amplifications, respectively.
Statistical analysis
All analyses were conducted in R, version 3.3.3 (http://www.R-project.org) and SPSS 22.0 (SPSS Inc., Chicago, IL, USA). Fisher’s exact tests and t-tests were performed. A two-sided P value of ≤0.05 was considered to show a statistically significant difference.
Results
Patient characteristics
The cohort was comprised of 67% males and 33% females with a median age of 38.5 years. Of the 36 ALCL patients evaluated by IHC, 14 were ALK-positive and 22 were ALK-negative. In three of the ALK-negative patients, DUSP22 rearrangement was detected by FISH. Fifteen patients displayed extranodal involvement at diagnosis, and two patients had skin involvement during their disease course. Overall, 50% (n=18) of patients received a CHOP/CHOP-like regimen as the first-line treatment. A 15-year-old patient and a 13-year-old patient received BFM90 and BFM95 regimens, respectively. One patient underwent radiotherapy. Five patients did not receive any treatment, and the remaining 10 patients had no treatment information available. The median follow-up time was 26 months and the median overall survival was 42 months (range, 1–103 months). Demographic and clinical characteristics of the patients are shown in Table 1.
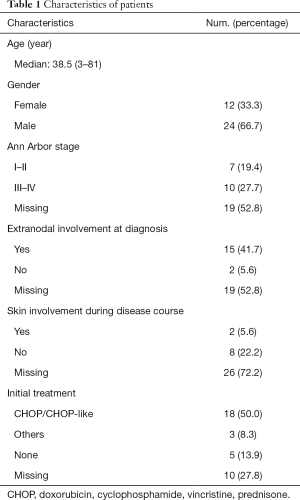
Full table
The mutation landscape of the entire cohort
Thirty-six patients with available baseline tumor tissue samples were included in the baseline mutation landscape analysis. A total of 102 mutations were identified from the entire cohort. The mutation landscape of the cohort is presented in Figure 1A. Mutations were detected in the tumor tissue samples of 81% (29/36) of patients. Twelve patients (33%) had ALK rearrangement, and one patient had an SNV of ALK. With IHC used as a reference, our capture-based targeted NGS had a sensitivity of 85.7% (12/14) and a specificity of 100% for identifying ALK status in biopsy samples. Two patients with ALK fusion detected by IHC were not identified by NGS. The tumor cell fraction in one of these samples was <30%, while the DNA from the other sample was heavily degraded when NGS was performed. Among the 12 patients who had ALK rearrangements revealed by NGS, nucleophosmin 1 (NPM1) was the most common gene fusion partner, which was harbored by 7 (58%) patients. The other ALK gene fusion partners detected from the cohort included 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (ATIC)-ALK (n=2), trafficking from ER to Golgi regulator (TFG) (n=1), tropomyosin 3 (TPM3) (n=1), and myosin heavy chain 9 (MYH9) (n=1). Other commonly seen mutations included tumor protein p53 (TP53) (19.4%) and lysine methyltransferase 2D (KMT2D) (11.1%).
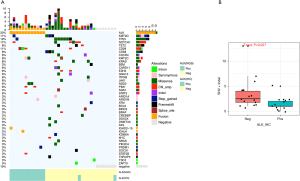
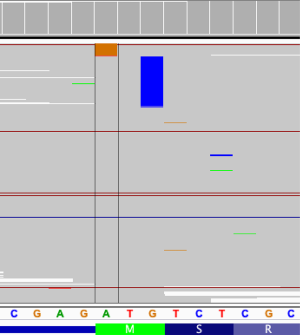
Compared with ALK-positive ALCLs, which have been well characterized at the molecular level, little is known about the genetic features of ALK-negative ALCLs. We compared the mutation landscape between patients with IHC-based ALK-negative and ALK-positive status. Patients with IHC-based ALK-negative status had significantly more SNVs and INDELs (P=0.027) (Figure 1B). Among the 22 IHC-based ALK-negative patients, the most common mutation was TP53 (n=6, 27.3%), followed by notch receptor 1 (NOTCH1) (n=5, 22.7%), KMT2D (n=3, 13.6%), KRAS proto-oncogene, GTPase (KRAS) (n=3, 13.6%), TET methylcytosine dioxygenase 2 (TET2) (n=3, 13.6%), Janus kinase 1 (JAK1) (n=2, 9.1%), signal transducer and activator of transcription 3 (STAT3) (n=1, 4.5%), and Fas cell surface death receptor (FAS) (n=1, 4.5%). Of note, start-loss of beta-2-microglobulin (B2M) was detected in a 53-year-old ALK-negative patient (Figure 2) who was diagnosed with stage IIA ALCL with liver involvement at diagnosis and had no skin involvement during the disease course. This patient received a CHOP-like regimen as first-line treatment and achieved partial response, with progression-free survival and OS of 13 months and 93 months and counting, respectively. DUSP22-rearrangement, a favorable prognostic biomarker of ALK-negative ALCL, was detected in the same patient.
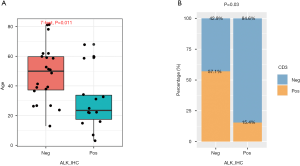
The correlation between ALK fusion status and various clinical parameters
The correlations between ALK fusion status and various clinical parameters were further investigated. ALK-positive ALCL was observed to be more common among younger patients (P=0.011), and ALK-negative patients were more likely to be CD3 positive (P=0.03) (Figure 3). No differences were observed between ALK fusion and Ki67 (P=0.64), sex (P=1), BCL2 (P=0.13), BCL6 (P=0.14), or disease stage (P=0.75).
Discussion
In this study, we revealed the somatic genomic landscapes of systemic ALCLs in 36 Chinese patients. We found that younger patients were more likely to be ALK positive, while patients with ALK wild-type (WT) ALCLs were more likely to have SNVs and INDELs than patients with ALK rearrangements. Overall, our study revealed new evidence of genetic heterogeneity between Chinese ALCL patients.
Due to the rarity of ALCL, the genetic and clinical heterogeneity of patients with the disease are not well understood. ALK-fusion is known to be more likely to occur in younger ALCL patients (4,7). The majority of ALK-rearranged ALCLs have translocations involving the ALK gene and its partner NPM1 (21). In the current cohort, the median age of the patients with ALK-positive ALCL was significantly younger than that of the ALK-negative patients. NPM1-ALK was the most common ALK rearrangement, which is consistent with the results of a previous study (21).
Compared with ALK-positive ALCLs, which are well characterized clinically, the genetic features of ALK-negative ALCLs are not well defined. Previous studies reported that the most commonly mutated genes in ALK-negative ALCL were JAK1, STAT3, PR/SET domain 1 (PRDM1), TP53, TET2, and FAS (22,23). The most common mutation among ALK-negative patients in our study was TP53 (27.3%), followed by NOTCH1 (22.7%), KMT2D (13.6%), KRAS (13.6%), TET2 (13.6%), JAK1 (9.1%), STAT3 (4.5%), and FAS (4.5%). However, PRDM1, which has been reported as a common mutation in ALK-negative ALCL patients, was not detected in our ALK-negative cohort, which indicates that Chinese systemic ALCL patients may have a unique somatic mutational profile. Recently, chromosomal rearrangements of the DUSP22 and TP63 genes were reported in ALK-negative ALCL, with an incidence of 30% and 8%, respectively (10,24). In our study, however, DUSP22 rearrangement was detected in three patients, accounting for only 13.6% of the ALK-negative ALCL patients; this discrepancy can be partly attributed to the small sample size. Detection of TP63 rearrangement was not performed in our cohort.
In our study, start-loss of B2M gene was detected in one ALK-negative patient. This patient showed a favorable prognosis, with OS of 93 months and counting. Inactivating B2M mutations, including exon-1 splice-donor, start codon mutations, out-of-frame first-exon deletions, and acceptor-site mutations, may induce the loss of expression of the major histocompatibility complex class I complex, which is vital for antigen presentation (25). Inactivating B2M mutations have been detected in both Hodgkin and non-Hodgkin lymphoma patients. A lack of B2M expression is associated with a favorable prognosis in Hodgkin lymphoma patients; in contrast, it is associated with unfavorable prognosis in non-Hodgkin lymphoma patients (25,26). Low B2M (≤3 mg/dL) has also been reported to be a favorable prognostic factor in ALCL (27). To the best of our knowledge, our study provides the first evidence of start-loss of B2M in an ALCL patient, thus suggesting the prognostic value of B2M in ALCL. This patient harbored DUSP22 rearrangement, which is a favorable prognostic biomarker of ALCL. Therefore, the prognostic value of B2M mutation needs to be further explored.
This study has some limitations. First, this is a retrospective study, which has all the drawbacks associated with a retrospective study including missing clinical data. Second, this study has a relatively small sample size, and limited clinical and follow-up data, which may have impacted the analyses. Third, this study was conducted in a single center that could introduce sample or population bias. Nevertheless, our study revealed the unique genomic profiles of Chinese systemic ALCL patients and represents an incremental step in improving the understanding of genetic heterogeneity in ALCL.
Acknowledgments
We would like to thank Mr. Bing Li, Dr. Han Han-Zhang, Dr. Lu Zhang, Dr. Xuejing Li, Dr. Analyn Lizaso, Dr. Ting Bei and Mr. Chanhe Li of Burning Rock Biotech for their help in data analysis, manuscript editing, and valuable discussion.
Funding: This research was supported by the Science and Technology Program of Fujian Province, China (No. 2018Y2003, No. 2018-CX-10, No. 2019L3018, and No. 2019Y91010045).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7574
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-7574
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7574). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures involving human participants were performed in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol was approved by the Ethics Committee of Fujian Medical University Cancer Hospital and Fujian Cancer Hospital (approval number: 2017-055-01). Written informed consent was provided by all the patients included in the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Montes-Mojarro IA, Steinhilber J, Bonzheim I, et al. The Pathological Spectrum of Systemic Anaplastic Large Cell Lymphoma (ALCL). Cancers (Basel) 2018;10:107. [Crossref] [PubMed]
- Hapgood G, Savage KJ. The biology and management of systemic anaplastic large cell lymphoma. Blood 2015;126:17-25. [Crossref] [PubMed]
- Eyre TA, Khan D, Hall GW, et al. Anaplastic lymphoma kinase-positive anaplastic large cell lymphoma: current and future perspectives in adult and paediatric disease. Eur J Haematol 2014;93:455-68. [Crossref] [PubMed]
- Gascoyne RD, Aoun P, Wu D, et al. Prognostic significance of anaplastic lymphoma kinase (ALK) protein expression in adults with anaplastic large cell lymphoma. Blood 1999;93:3913-21. [Crossref] [PubMed]
- Katayama R, Lovly CM, Shaw AT. Therapeutic targeting of anaplastic lymphoma kinase in lung cancer: a paradigm for precision cancer medicine. Clin Cancer Res 2015;21:2227-35. [Crossref] [PubMed]
- Fisher RI, Gaynor ER, Dahlberg S, et al. Comparison of a standard regimen (CHOP) with three intensive chemotherapy regimens for advanced non-Hodgkin's lymphoma. N Engl J Med 1993;328:1002-6. [Crossref] [PubMed]
- Savage KJ, Harris NL, Vose JM, et al. ALK- anaplastic large-cell lymphoma is clinically and immunophenotypically different from both ALK+ ALCL and peripheral T-cell lymphoma, not otherwise specified: report from the International Peripheral T-Cell Lymphoma Project. Blood 2008;111:5496-504. [Crossref] [PubMed]
- d'Amore F, Relander T, Lauritzsen GF, et al. Up-front autologous stem-cell transplantation in peripheral T-cell lymphoma: NLG-T-01. J Clin Oncol 2012;30:3093-9. [Crossref] [PubMed]
- Onaindia A, de Villambrosia SG, Prieto-Torres L, et al. DUSP22-rearranged anaplastic lymphomas are characterized by specific morphological features and a lack of cytotoxic and JAK/STAT surrogate markers. Haematologica 2019;104:e158-62. [Crossref] [PubMed]
- Parrilla Castellar ER, Jaffe ES, Said JW, et al. ALK-negative anaplastic large cell lymphoma is a genetically heterogeneous disease with widely disparate clinical outcomes. Blood 2014;124:1473-80. [Crossref] [PubMed]
- Pedersen MB, Hamilton-Dutoit SJ, Bendix K, et al. DUSP22 and TP63 rearrangements predict outcome of ALK-negative anaplastic large cell lymphoma: a Danish cohort study. Blood 2017;130:554-7. [Crossref] [PubMed]
- Hapgood G, Ben-Neriah S, Mottok A, et al. Identification of high-risk DUSP22-rearranged ALK-negative anaplastic large cell lymphoma. Br J Haematol 2019;186:e28-e31. [PubMed]
- NCCN Clinical Practice Guidelines in Oncology for T-cell lymphomas. Version 3. 2020 [database on the Internet]. National Comprehensive Cancer Network (NCCN). 2020. Available online: https://www.nccn.org/. Accessed: June 15, 2020.
- Younes A, Hilden P, Coiffier B, et al. International Working Group consensus response evaluation criteria in lymphoma (RECIL 2017). Annals of Oncology 2017;28:1436-47. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. [Crossref] [PubMed]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. [Crossref] [PubMed]
- Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568-76. [Crossref] [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [Crossref] [PubMed]
- Cingolani P, Platts A. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80-92. [Crossref] [PubMed]
- Newman AM, Bratman SV, Stehr H, et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics 2014;30:3390-3. [Crossref] [PubMed]
- Morris SW, Kirstein MN, Valentine MB, et al. Sequence Correction. Science 1995;267:316-7. [Crossref] [PubMed]
- Crescenzo R, Abate F, Lasorsa E, et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell 2015;27:516-32. [Crossref] [PubMed]
- Mereu E, Pellegrino E, Scarfo I, et al. The heterogeneous landscape of ALK negative ALCL. Oncotarget 2017;8:18525-36. [Crossref] [PubMed]
- Feldman AL, Dogan A, Smith DI, et al. Discovery of recurrent t(6;7)(p25.3;q32.3) translocations in ALK-negative anaplastic large cell lymphomas by massively parallel genomic sequencing. Blood 2011;117:915-9. [Crossref] [PubMed]
- Reichel J, Chadburn A, Rubinstein PG, et al. Flow sorting and exome sequencing reveal the oncogenome of primary Hodgkin and Reed-Sternberg cells. Blood 2015;125:1061-72. [Crossref] [PubMed]
- Amiot L, Onno M, Lamy T, et al. Loss of HLA molecules in B lymphomas is associated with an aggressive clinical course. Br J Haematol 1998;100:655-63. [Crossref] [PubMed]
- Sibon D, Fournier M, Briere J, et al. Long-term outcome of adults with systemic anaplastic large-cell lymphoma treated within the Groupe d'Etude des Lymphomes de l'Adulte trials. J Clin Oncol 2012;30:3939-46. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)


