An aberrant DNA methylation signature for predicting hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC), which is one of the main types of liver cancer, is the fourth deadliest cancer worldwide. According to global cancer statistics, in recent years, HCC incidence has risen by approximately 3% annually (1). Because HCC patients are often diagnosed at an advanced stage, the disease has extremely poor survival rates and prognosis. Many factors contribute to the progression of HCC, including nonalcoholic fatty liver disease, alcohol addiction, and hepatitis C virus (2,3). The initial tumorigenesis and progression of HCC are complex and involve many factors. However, the exact molecular mechanism responsible for the poor prognosis of HCC patients is still unknown. So far, advances in liver cancer treatments, such as liver transplantation, surgical resection, and radiofrequency ablation, have failed to improve the dismal 5-year survival rate of patients, which stands at less than 20% (4,5). The poor prognosis of HCC mostly results from the limited understanding of the molecular factors associated with the disease and the lack of effective interventions (6,7). Like vascular endothelial growth factor (VEGF) inhibitors and mammalian target of rapamycin (mTOR) inhibitors have been investigated according to the knowledge about some molecular domain (8). Although these molecular sites have been investigated as potential therapeutic targets in HCC, few have shown promise (9,10). Therefore, there is an urgent need to identify new biomarkers for prognostic prediction and treatment in patients with HCC.
According to previous research, the occurrence and progression of cancer is mainly related to the expression or structural alteration of tumor-associated genes. Recent studies have demonstrated that DNA methylation of tumor-related genes also plays a crucial role in tumor development (11,12). Scientists believe that DNA methylation is often involved in the mechanism of gene silencing and plays an important role in a number of biological processes, including development of embryo, transcription of DNA, imprint of genome, and inactivation of X-chromosome (13-15). Signatures derived from the methylation of DNA have already been studied in some cancers. Methylation of the CDH1 promoter, for instance, can indicate poor prognosis in patients with breast cancer (16), while in patients with stage I non-small-cell lung cancer, methylation of RUSSF1A, P16, CDH13, and APC is predictive of a higher rate of recurrence (17,18). In the study of HCC, several methylation biomarkers have also been used to predict prognosis (19). For example, CELSR3 (20), SEPT9 (21), TNFRSF12A (22), TP73 (23), RUNX3, and RASSF1A are single methylation biomarkers for predicting prognosis (24). However, many genes are involved in tumorigenesis, which makes some of them unsuitable as biomarkers (25). Although the mechanisms underlying the occurrence and development of HCC are extremely complex, it would be meaningful to establish a molecular biomarker model with good ability to discriminate the prognosis of patients with HCC.
In epigenetic research, methylation may be a symbolic event during carcinogenesis. Several methylations, such as promoter methylation and CpG-island methylation, are closely associated with HCC. Therefore, different from previous studies, we aimed to identify survival-related DNA methylation signatures in HCC to provide potential therapeutic targets by using bioinformatic methods. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7804).
Methods
Profiling DNA methylation and gene expression datasets of HCC patients
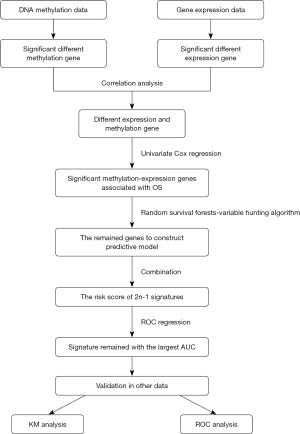
We extracted a gene expression dataset (Illumina HiSeq RNA Seq V2) from the The Cancer Genome Atlas (TCGA) database. The DNA methylation profiles (Illumina Human Methylation 450K Bead Chip Array) of samples from 392 HCC patients and 49 healthy volunteers were also extracted. Clinical information of patients in the gene expression and methylation datasets, including survival time, health status, sex, age and clinical tumor stage, are shown in Table 1. To improve the accuracy of the data, we preprocessed the datasets, which included removing the sites for which 70% methylated level was not available and removing genes if their expression was not detected in >30% of patients. Genes with an RPKM expression of 0 in all samples were also excluded (26). A flowchart of this study is shown in Figure 1. This study was a bioinformatic analysis based on data from an open database, which involved no ethic problem.

Full table
Identification of DMGs and DEGs associated with HCC
In order to find the differentially expressed methylated genes, the Benjamini-Hochberg false-discovery rate (FDR) method was adopted to adjust the P value. DMGs were named as those with a fold change >2 with a P value <0.05, an FDR <0.05, and a beta value >0.1. DEGs met the criteria of a fold change >2 with a P value <0.05, an FDR <0.05, and an FPKM >1.
With the identification of DMGs and DEGs, we investigated 37 hub genes that were differentially expressed and enriched in different methylated genes between the DMGs and DEGs.
Construction of DNA methylation signature model
A predictive model was established using methods previously described (27,28). Methylation often occurs at specific loci of target genes and often involves many methylation sites. To enhance the accuracy of the detection results in this study, we only searched methylation sites of hub genes, and then identified which site was related to survival in patients with HCC. The HCC methylation samples were randomly divided into two groups at a ratio of 1:2: the test group (n=131) and the training group (n=261). Univariate Cox analysis was carried out to examine the relationship between survival time and health status and each methylation site in the training dataset (29). It was generally assumed that a model was more practical if its render had a smaller number of DNA methylation sites. Therefore, we used a random survival forests-variable hunting (RSFVH) algorithm to screen the gene sites. To determine the most powerful and accurate DNA methylation sites for predicting the prognosis of HCC in our study, multivariate Cox analysis was applied to create the prognostic model according to the following formula:
[1]
In which N means the number of methylation sites, Methi represents the value of methylation sites, and Coefi represents a univariate Cox regression coefficient. When the coefficient of the Coi was <0, we defined the site as a useful prognostic site, sites with a Coi coefficient of >0 were considered not to be used as prognostic sites. Risk score (RS) was the multi-node weighted sum of the risk scores.
Function of the selected DNA methylation signature genes
We performed Gene Ontology (GO) analysis (http://www.geneontology.org) to study the function of the selected DNA methylation signature genes.
Statistical analysis
A risk model was constructed incorporating the selected methylation sites. With this model, HCC patients were divided into high- and low-risk groups, with the median risk score used as a cutoff value in the training dataset. Kaplan-Meier survival curve analysis and receive operating characteristic (ROC) curve analysis were conducted to test the predictive value of the methylation gene site signatures. We further tested the predictive value of the DNA methylation signature in the test dataset. Then, multivariable Cox analysis was used to identify whether the DNA methylation signature was an independent risk factor for survival in HCC. P<0.05 was considered to show a statistically significant difference. Fisher’s exact test and χ2 tests were used to select significant GO and KEGG pathway categories, with the threshold for significance set at P<0.05. All analysis was conducted with the R statistical software (version 3.5.1, Mathsoft Inc., NY, USA).
Results
DMGs and DEGs associated with HCC
In the methylation dataset, we identified 203 DMGs (P<0.05; Δβ >0.1), with 23 hypomethylated genes and 180 hypermethylated genes. There were 4,880 DEGs identified, including 3,818 upregulated genes and 1,062 downregulated genes. Finally, a total of 37 hub genes were identified (Figure 2).

DNA methylation signature sites in the training group
A total of 713 methylation sites were identified for the 37 hub genes. The univariate Cox proportional hazards regression analysis revealed that 36 methylated sites were associated with survival of HCC patients (P<0.05, Figure 3A). Then, an RSFVH was employed to determine which of the sites were DNA methylation signature sites. Ten sites related to the patient survival were selected by the RSFVH algorithm according to the permutation importance score (PFI). To select the signature with the greatest predictive power, a multivariable Cox regression analysis was conducted, and a model was built with seven methylated gene sites set (cg12824782, cg24871714, cg18683774, cg22796509, cg19450025, cg10474350, and cg06511917, Figure 3B) to assess the survival risk of the HCC patients. Risk scores were determined as follows (higher RS predicts higher risk of death):

[2]
Notes: RS = risk score; meth = methylation value.
Power of the DNA methylation signature to predict survival in patients with HCC
Each patient was assigned a risk score from the model derived from the selected methylated sites, and the median value of risk score was used as the cut-off point to divide the patients in the training group into group of low-risk (LRG, n=131) or group of high-risk (HRG, n=130). The KM survival analysis demonstrated that compared with HRG, the overall survival (OS) in LRG was significantly higher (OS: 2.81 vs. 2.11 years; log-rank test, P<0.05; Figure 4A). The predictive power of the model was validated in the test group, and a significant difference was again found between the HRG and LRG. In the test group, the OS of the high-risk patients was significantly lower than that of the low-risk patients (OS: 2.04 vs. 2.88 years; log-rank test, P<0.05; Figure 4B).

Further, the ROC analysis revealed the seven methylation signature sites to have a high predictive ability in the training group (AUC Signature =0.750, Figure 4C), which demonstrated a good performance of our model in predicting the survival of patients with HCC (AUC Signature =0.742, Figure 4D). Multivariable Cox regression analysis from the training dataset showed that after adjustment for clinical features including sex, age, and pTNM, the signature risk score was an independent prognostic factor of HCC (Figure 5A).

Functional annotation of DNA methylated genes signature
GO functional annotation analysis showed that the DNA methylation genes were significantly enriched in DNA-binding transcription activator activity, RNA polymerase II distal enhancer sequence-specific DNA binding, and enhancer sequence-specific DNA binding (P<0.05). Therefore, the seven gene sites may participate in the tumorigenesis through regulating cellular metabolic processes (Figure 5B).
Discussion
For doctors and investigators, HCC remains a clinical challenge for multiple reasons, including delayed diagnosis (with many HCC patients diagnosed at stage 4, leaving them with no effective treatment options), histologic heterogeneity, poor management results, and limited treatment choices (30). However, the mechanism of HCC carcinogenesis is extremely complicated and has yet to be fully illuminated. Gene mutation, immune escape, protein expression disorders, and blood vessel malformation are all involved in cancers. As gene mutation is the main and original mechanism of cancer occurrence, the majority of previous studies have focused on genes that are already recognized as being associated with HCC. Traditionally, HCC prognosis has been determined based on the TNM staging system and other baseline characteristics, including blood test results, history of disease, and age. However, clinical outcomes differ greatly, even among patients at the same stage. Therefore, due to the unpredictable clinical outcomes among patients, the TNM staging system is insufficient for guiding personalized treatment (31). In the management of cancer patients, precise medicine has made significant progress. Molecular targeted drugs can achieve a good prognosis in many malignancies, including breast, stomach, colon, and liver cancer. However, despite this progress, some patients survive for only a short time after diagnosis.
In recent years, several powerful predictive biomarkers have been reported to improve the clinical management of cancer. DNA methylation is a well-known epigenetic change which can alter the expression of vital tumorigenesis-associated genes without changing the genetic sequence (32). These changes have been shown to be closely associated with the occurrence and development of cancer, and many related biomarkers have been reported (33). Tumor-specific methylation sites are critical for the inchoate diagnosis of cancer (34). Since aberrant DNA methylation has been proven to be associated with the tumorigenesis of HCC, it may serve as a predictive biomarker for the prognosis of HCC patients (35,36). However, epigenetic biomarkers for HCC have rarely been investigated, with most research on prognostic biomarkers of HCC focused on messenger RNAs (mRNAs), long non-coding RNAs, and microRNAs (miRNAs). For instance, Ma et al. reported that LINC02499 performed well in the diagnosis of HCC, with a sensitivity and specificity of 83.8%. Moreover, the up-regulation and down-regulation of LINC02499 was observed to significantly affect the proliferation, migration, and invasion of HCC cells in vitro. Therefore, LINC02499 may be a useful predictive biomarker in the diagnosis and prognosis of HCC (37). Wang et al. found that up-regulation of miR-3174 inhibited cell apoptosis and promote cell proliferation in HCC by down-regulating the expression of FOXO1, suggesting that miR-3174 may be a new therapeutic candidate for HCC patients (38). However, although some prognostic biomarkers of HCC have been identified, the results of studies so far have been inconsistent; moreover, an analysis at multiple omics levels has yet to be performed. Therefore, a reliable molecular model to predict the survival of HCC patients more precisely needs to be developed and validated. In the present study, we obtained 713 methylated gene sites from 37 hub genes. Using various statistical methods, we identified 7 methylation signature sites from the methylated sites. The signature that we established was able to differentiate HCC patients into high-risk and low-risk groups with significantly different survival duration, both in the training and test datasets, indicating that it has a satisfying predictive power. The DNA methylation site signature was also found to be independent of other clinical characteristics. Moreover, we found that the methylation gene site signature was better than other clinical risk factors, such as stage and age, in predicting OS. Therefore, our findings contribute to the understanding of the development of HCC as well as the development of targeted therapies that may ultimately improve the survival of HCC patients. Experimental studies demonstrated that when the methylation level is reduced, it can cause gene activation and tumor occurrence. Accordingly, investigators assumed that DNA methylation has a strong relationship with tumor development and prognosis.
Additionally, we conducted a functional analysis of the selected DNA methylation genes. The seven methylation sites were located in ZNF382, ZNF529, IRX5, PTF1A, ZIC1, SLX1A, and HOXA7, respectively. GO function annotation analysis showed that the genes were significantly enriched in DNA-binding transcription activator activity, RNA polymerase II distal enhancer sequence-specific DNA binding, and enhancer sequence-specific DNA binding. Zinc-finger protein 382 (ZNF382), which encodes a putative 64 kDa zinc finger protein, is located on chromosome 19q13.13 (39). It has been reported that ZNF382 may play a significant tumor-suppressive role and methylate in multiple cancers, including colon, esophageal, breast, nasopharyngeal, and gastric cancer (40,41). Dang et al. reported that ZNF382 inhibited HCC tumorigenesis by impairing the activity of AP-1 and the activation of the p53 pathway (42). Iroquois homeobox (IRX) genes also play a key role in the development of malignancies. IRX5 is a transcription factor that has a variety of functions in different cancers (43). As a key transcription factor, IRX5 regulates the key regulatory genes to regulate invasion, cell growth, apoptosis, and migration, thus promoting the occurrence and development of many tumors. Recent data indicate that IRX5 significantly promotes tumor growth in tongue squamous cell carcinoma by targeting the OPN promoter and activating the NF-κB pathway (44). In HCC, IRX5 overexpression can promote cell proliferation and tumorigenesis (45), while knockdown of IRX5 can affect cell apoptosis via the p53 pathway (46). ZIC1 is a transcriptional cofactor (47,48), and there is growing evidence that it has an antitumor effect in a variety of epithelial cancers, including colon, endometrial, breast, and thyroid cancers (49-52). Ge et al. showed that ZIC1 inhibits GC transfer by attenuating the EMT process and Wnt/β-catenin pathway (53,54). HOXA7, meanwhile, plays important roles in cell differentiation and cell morphological development. In a recent study, for instance, it was shown to play a key role in normal cell proliferation and differentiation, and its overexpression can lead to the development of malignant tumors (55). Significantly increased HOXA7 expression has also been found in ovarian (56), breast (57), lung (58), and gastric cancer (59). However, ZNF529, PTF1A, and SLX1A are less well known, having been the focus of few articles (60,61). The results of the current study provide a novel direction for further research of the potential prognostic role of these genes in HCC.
Unfortunately, this study has some limitations that need to be highlighted. Firstly, this is a retrospective study, and it is likely that inherent biases influenced the final results to some degree. Hence, we may have missed some signatures that are potentially correlated with survival in patients with HCC. Secondly, we did not further investigate the mechanism of the DNA methylation genes in the occurrence and development of HCC. Finally, further studies with larger sample sizes and longer duration of follow-up are needed to validate our findings.
Acknowledgments
Funding: New academic talent of Guizhou Medical University (19NSP012).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7804
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7804). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was a bioinformatic analysis based on data from an open database, which involved no ethic problem.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Singal AG, El-Serag HB. Hepatocellular carcinoma from epidemiology to prevention: translating knowledge into practice. Clin Gastroenterol Hepatol 2015;13:2140-51. [Crossref] [PubMed]
- McGlynn KA, Petrick JL, London WT. Global epidemiology of hepatocellular carcinoma: an emphasis on demographic and regional variability. Clin Liver Dis 2015;19:223-38. [Crossref] [PubMed]
- Cheung TT. Management of hepatocellular carcinoma: from bench to bedside and beyond. Transl Gastroenterol Hepatol 2019;4:54. [Crossref] [PubMed]
- Kudo M. Systemic therapy for hepatocellular carcinoma: latest advances. Cancers (Basel) 2018;10:412. [Crossref] [PubMed]
- Li D, Liu X, Zhou J, et al. Long noncoding RNA HULC modulates the phosphorylation of YB-1 through serving as a scaffold of extracellular signal-regulated kinase and YB-1 to enhance hepatocarcinogenesis. Hepatology 2017;65:1612-27. [Crossref] [PubMed]
- Ghanem I, Riveiro ME, Paradis V, et al. Insights on the CXCL12-CXCR4 axis in hepatocellular carcinoma carcinogenesis. Am J Transl Res 2014;6:340-52. [PubMed]
- Montironi R, Cheng L, Scarpelli M, et al. Pathology and genetics: tumours of the urinary system and male genital system: clinical implications of the 4th edition of the who classification and beyond. Eur Urol 2016;70:120-3.
- Pratama MY, Pascut D, Massi MN, et al. The role of microRNA in the resistance to treatment of hepatocellular carcinoma. Ann Transl Med 2019;7:577. [Crossref] [PubMed]
- Huang Z, Zhou JK, Peng Y, et al. The role of long noncoding RNAs in hepatocellular carcinoma. Mol Cancer 2020;19:77. [Crossref] [PubMed]
- Lorincz AT. Cancer diagnostic classifiers based on quantitative DNA methylation. Expert Rev Mol Diagn 2014;14:293-305. [Crossref] [PubMed]
- Plass C, Pfister SM, Lindroth AM, et al. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet 2013;14:765-80. [Crossref] [PubMed]
- Guo H, Zhu P, Yan L, et al. The DNA methylation landscape of human early embryos. Nature 2014;511:606-10. [Crossref] [PubMed]
- Robertson KD. DNA methylation and human disease. Nat Rev Genet 2005;6:597-610. [Crossref] [PubMed]
- Xu X. DNA methylation and cognitive aging. Oncotarget 2015;6:13922-32. [Crossref] [PubMed]
- Liu J, Sun X, Qin S, et al. CDH1 promoter methylation correlates with decreased gene expression and poor prognosis in patients with breast cancer. Oncol Lett 2016;11:2635-43. [Crossref] [PubMed]
- Kim-Wanner SZ, Assenov Y, Nair MB, et al. Genome-wide DNA methylation profiling in early stage I lung adenocarcinoma reveals predictive aberrant methylation in the promoter region of the long noncoding RNA plut: an exploratory study. J Thorac Oncol 2020;15:1338-50. [Crossref] [PubMed]
- Sun F, Li L, Yan P, et al. Causative role of PDLIM2 epigenetic repression in lung cancer and therapeutic resistance. Nat Commun 2019;10:5324. [Crossref] [PubMed]
- Hua S, Ji Z, Quan Y, et al. Identification of hub genes in hepatocellular carcinoma using integrated bioinformatic analysis. Aging (Albany NY) 2020;12:5439-5468. [Crossref] [PubMed]
- Xie Z, Dang Y, Wu H, et al. Effect of CELSR3 on the cell cycle and apoptosis of hepatocellular carcinoma cells. J Cancer 2020;11:2830-44. [Crossref] [PubMed]
- Kotoh Y, Suehiro Y, Saeki I, et al. Novel liquid biopsy test based on a sensitive methylated SEPT9 assay for diagnosing hepatocellular carcinoma. Hepatol Commun 2020;4:461-70. [Crossref] [PubMed]
- Wang Y, Zhang S, Xie X, et al. Association of TNFRSF12A methylation with prognosis in hepatocellular carcinoma with history of alcohol consumption. Front Genet 2020;10:1299. [Crossref] [PubMed]
- Yao Z, Di Poto C, Mavodza G, et al. DNA methylation activates TP73 Expression in hepatocellular carcinoma and gastrointestinal cancer. Sci Rep 2019;9:19367. [Crossref] [PubMed]
- El-Bendary M, Nour D, Arafa M, et al. Methylation of tumour suppressor genes RUNX3, RASSF1A and E-Cadherin in HCV-related liver cirrhosis and hepatocellular carcinoma. Br J Biomed Sci 2020;77:35-40. [Crossref] [PubMed]
- Brooks JD. Translational genomics: the challenge of developing cancer biomarkers. Genome Res 2012;22:183-7. [Crossref] [PubMed]
- Xu J, Li Y, Lu J, et al. The mRNA related ceRNA-ceRNA landscape and significance across 20 major cancer types. Nucleic Acids Res 2015;43:8169-82. [Crossref] [PubMed]
- Hu S, Yin X, Zhang G, et al. Identification of DNA methylation signature to predict prognosis in gastric adenocarcinoma. J Cell Biochem 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Guo JC, Wu Y, Chen Y, et al. Protein-coding genes combined with long noncoding RNA as a novel transcriptome molecular staging model to predict the survival of patients with esophageal squamous cell carcinoma. Cancer Commun (Lond) 2018;38:4. [Crossref] [PubMed]
- Kumar A. Letter to the editor: Role of sorafenib in liver transplantation outcomes of patients with hepatocellular carcinoma presenting beyond milan criteria? Hepatology 2020. [Epub ahead of print].
- Jiang W, Liu N, Chen XZ, et al. Genome-wide identification of a methylation gene panel as a prognostic biomarker in nasopharyngeal carcinoma. Mol Cancer Ther 2015;14:2864-73. [Crossref] [PubMed]
- Peters FS, Manintveld OC, Betjes MG, et al. Clinical potential of DNA methylation in organ transplantation. J Heart Lung Transplant 2016;35:843-50. [Crossref] [PubMed]
- Higgins-Chen AT, Boks MP, Vinkers CH, et al. Schizophrenia and epigenetic aging biomarkers: increased mortality, reduced cancer risk, and unique clozapine effects. Biol Psychiatry 2020;88:224-35. [Crossref] [PubMed]
- Ying J, Xu T, Wang Q, et al. Exploration of DNA methylation markers for diagnosis and prognosis of patients with endometrial cancer. Epigenetics 2018;13:490-504. [Crossref] [PubMed]
- Warton K, Mahon KL, Samimi G. Methylated circulating tumor DNA in blood: power in cancer prognosis and response. Endocr Relat Cancer 2016;23:R157-71. [Crossref] [PubMed]
- Wu X, Li J, Gassa A, et al. Circulating tumor DNA as an emerging liquid biopsy biomarker for early diagnosis and therapeutic monitoring in hepatocellular carcinoma. Int J Biol Sci 2020;16:1551-62. [Crossref] [PubMed]
- Huang X, Yang C, Wang J, et al. Integrative analysis of DNA methylation and gene expression reveals distinct hepatocellular carcinoma subtypes with therapeutic implications. Aging (Albany NY) 2020;12:4970-95. [Crossref] [PubMed]
- Ma X, Mo M, Tan HJJ, et al. LINC02499, a novel liver-specific long non-coding RNA with potential diagnostic and prognostic value, inhibits hepatocellular carcinoma cell proliferation, migration, and invasion. Hepatol Res 2020;50:726-40. [Crossref] [PubMed]
- Wang Q, Yang X, Zhou X, et al. MiR-3174 promotes proliferation and inhibits apoptosis by targeting FOXO1 in hepatocellular carcinoma. Biochem Biophys Res Commun 2020;526:889-97. [Crossref] [PubMed]
- Luo K, Yuan W, Zhu C, et al. Expression of a novel Krüpple-like zinc-finger gene, ZNF382, in human heart. Biochem Biophys Res Commun 2002;299:606-12. [Crossref] [PubMed]
- Cheng Y, Geng H, Cheng SH, et al. KRAB zinc finger protein ZNF382 is a proapoptotic tumor suppressor that represses multiple oncogenes and is commonly silenced in multiple carcinomas. Cancer Res 2010;70:6516-26. [Crossref] [PubMed]
- Zhang C, Xiang T, Li S, et al. The novel 19q13 KRAB zinc-finger tumour suppressor ZNF382 is frequently methylated in oesophageal squamous cell carcinoma and antagonises Wnt/β-catenin signalling. Cell Death Dis 2018;9:573. [Crossref] [PubMed]
- Dang S, Zhou J, Chen Y, et al. Dynamic expression of ZNF382 and its tumor-suppressor role in hepatitis B virus-related hepatocellular carcinogenesis. Oncogene 2019;38:4804-19. [Crossref] [PubMed]
- Sun X, Jiang X, Wu J, et al. IRX5 prompts genomic instability in colorectal cancer cells. J Cell Biochem 2020;121:4680-9. [Crossref] [PubMed]
- Zhu Q, Wu Y, Yang M, et al. IRX5 promotes colorectal cancer metastasis by negatively regulating the core components of the RHOA pathway. Mol Carcinog 2019;58:2065-76. [Crossref] [PubMed]
- Huang L, Song F, Sun H, et al. IRX5 promotes NF-κB signalling to increase proliferation, migration and invasion via OPN in tongue squamous cell carcinoma. J Cell Mol Med 2018;8:3899-910. [Crossref]
- Zhu L, Dai L, Yang N, et al. Transcription factorIRX5 promotes hepatocellular carcinoma proliferation and inhibits apoptosis by regulating the p53 signalling pathway. Cell Biochem Funct 2020;38:621-9. [Crossref] [PubMed]
- Aruga J, Minowa O, Yaginuma H, et al. Mouse Zic1 is involved in cerebellar development. J Neurosci 1998;18:284-93. [Crossref] [PubMed]
- Aruga J. The role of Zic genes in neural development. Mol Cell Neurosci 2004;26:205-21. [Crossref] [PubMed]
- Ali RG, Bellchambers HM, Arkell RM. Zinc fingers of the cerebellum (Zic): transcription factors and co-factors. Int J Biochem Cell Biol 2012;44:2065-8. [Crossref] [PubMed]
- Wong YF, Cheung TH, Lo KW, et al. Identification of molecular markers and signaling pathway in endometrial cancer in Hong Kong Chinese women by genome-wide gene expression profiling. Oncogene 2007;26:1971-82. [Crossref] [PubMed]
- Gan L, Chen S, Zhong J, et al. ZIC1 is downregulated through promoter hypermethylation, and functions as a tumor suppressor gene in colorectal cancer. PLoS One 2011;6:e16916. [Crossref] [PubMed]
- Qiang W, Zhao Y, Yang Q, et al. ZIC1 is a putative tumor suppressor in thyroid cancer by modulating major signaling pathways and transcription factor FOXO3a. J Clin Endocrinol Metab 2014;99:E1163-72. [Crossref] [PubMed]
- Han W, Cao F, Gao XJ, et al. ZIC1 acts a tumor suppressor in breast cancer by targeting survivin. Int J Oncol 2018;53:937-48. [Crossref] [PubMed]
- Ge Q, Hu Y, He J, et al. Zic1 suppresses gastric cancer metastasis by regulating Wnt/β-catenin signaling and epithelial-mesenchymal transition. FASEB J 2020;34:2161-72. [Crossref] [PubMed]
- Gomes AR, Zhao F, Lam EW. Role and regulation of the forkhead transcription factors FOXO3a and FOXM1 in carcinogenesis and drug resistance. Chin J Cancer 2013;32:365-70. [Crossref] [PubMed]
- Ota T, Gilks CB, Longacre T, et al. HOXA7 in epithelial ovarian cancer: interrelationships between differentiation and clinical features. Reprod Sci 2007;14:605-14. [Crossref] [PubMed]
- Zhang Y, Cheng JC, Huang HF, et al. Homeobox A7 stimulates breast cancer cell proliferation by up-regulating estrogen receptor-alpha. Biochem Biophys Res Commun 2013;440:652-7. [Crossref] [PubMed]
- Calvo R, West J, Franklin W, et al. Altered HOX and WNT7A expression in human lung cancer. Proc Natl Acad Sci U S A 2000;97:12776-81. [Crossref] [PubMed]
- Yang YC, Wang SW, Wu IC, et al. A tumorigenic homeobox (HOX) gene expressing human gastric cell line derived from putative gastric stem cell. Eur J Gastroenterol Hepatol 2009;21:1016-23. [Crossref] [PubMed]
- Medves S, Auchter M, Chambeau L, et al. A high rate of telomeric sister chromatid exchange occurs in chronic lymphocytic leukaemia B-cells. Br J Haematol 2016;174:57-70. [Crossref] [PubMed]
- Metzger P, Kirchleitner SV, Kluge M, et al. Immunostimulatory RNA leads to functional reprogramming of myeloid-derived suppressor cells in pancreatic cancer. J Immunother Cancer 2019;7:288. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)