
Kiaa0101 serves as a prognostic marker and promotes invasion by regulating p38/snail1 pathway in glioma
Introduction
Glioblastoma multiforme (GBM) is one of the most common malignant brain tumors, with an incidence range of 4.32–5.02 per 100,000 people in 2015 (1). To date, clinical prognosis of GBM patients remains unsatisfactory despite the wide use of current standard therapy regimens, which comprise maximum safe surgical resection followed by temozolomide and simultaneous radiotherapy, as well as emerging new treatments. Previous studies have reported a variation in survival rate among different races of patients with GBM, with 1- and 5-year rates found to be 41.4% and 5.4%, respectively across all population (2). The poor therapeutic effect and short-term recurrence have been attributed to infiltrative nature of GBM cells (3), which makes it difficult for complete resection to be obtained, because maximizing the extent of resection might bring additional complications and decrease quality of life in GBM patients (4). Numerous molecules play a crucial role in glioma tumorigenesis. Therefore, it is imperative to unravel the novel approaches that specifically target the complex gene network.
Kiaa0101, also known as p15 (PAF), is 15-KD protein containing a proliferating cell nuclear antigen (PCNA)-binding motif (5). Several studies have reported that this protein is overexpressed in many malignant tumors, including hepatocellular carcinoma, ovarian cancer and breast cancer (6,7). This expression has subsequently been associated with prognosis of patients with malignant tumors (8-10). A recent study demonstrated preferential Kiaa0101 overexpression in glioma stem cells. Additionally, knocking out Kiaa0101 not only impaired maintenance of self-renewal, but also reduced tumor-initiating cell frequency (11). Furthermore, Kiaa0101 knockdown was found to inhibit invasion of cancer cells by regulating epithelial-mesenchymal transition, which was accompanied by upregulation of E-cadherin as well as downregulation of N-cadherin, vimentin, TWIST1, ZEB1, and SNAIL. However, the mechanism by which Kiaa0101 affects migration and invasion of glioma cells remains unclear.
The mitogen-activated protein kinase (MAPK) signaling pathway, comprising a cascade of cascade-activated serine/threonine protein kinases, is a highly conserved signal transduction pathway (12). Functionally, MAPK plays a crucial role in regulating cell proliferation, cell cycle, invasion, migration and malignant transformation (13,14). Besides, the pathway is reportedly activated in over 88% of gliomas (15). Furthermore, several studies have closely associated its activation with the initiation and progression of glioma (16,17). Kiaa0101 was also shown to be a regulator of the MEK/ERK signaling pathway, where it plays a key role in mediating cell proliferation of hepatocellular carcinoma. However, the interaction between Kiaa0101 and MAPK in glioma remains unknown.
We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-3219).
Methods
Patients samples
Paraffin-embedded microarrays, comprising 97 glioma samples were used. Another 36 frozen samples, comprising 10 and 26 non-tumor brain and glioma samples, respectively, were analyzed using western blot (WB) and RT-PCR. All specimens were obtained from hospitalized patients at the department of neurosurgery of Renmin Hospital of Wuhan University, between January 2016 and March 2018. None of the patients received chemo- or radiotherapy before surgery. Upon collection, all brain tissues were immediately frozen and stored at −80 °C until biochemical assays.
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. The study was approved by the Institutional Ethics Committee of the Faculty of Medicine at Renmin Hospital of Wuhan University [approval number: 2012LKSZ (010) H], and all patients signed informed consents prior to inclusion.
Immunohistochemical staining
Glioma tissue microarrays were dewaxed and hydrated, then antigen retrieval conducted in a bottle full of antigen retrieval buffer (10 mmol/L, pH 6.0) for 15 min. Thereafter, we used 3% H2O2 to eliminate the endogenous peroxidase, then incubated the sections with primary anti-Kiaa0101 monoclonal antibody overnight at 4 °C. The sections were incubated with HRP-labeled polymer conjugated with secondary antibody for 30 min at 37 °C. The contents were stained using diaminobenzidine (DAB) for 2 min, followed by a second staining with hematoxylin. Images were captured using an Olympus BX40 microscope (Tokyo, Japan), with six high magnification fields randomly selected for observation. Kiaa0101 was mainly located in the nucleus and the positive staining cells appeared as brownish yellow grains. Final results were based on the strength of staining and the number of positive cells. The rate of positive staining (number of cells that were positive for staining per 100 cells) was scored as: 0 points for less than 5%, 1 point for 5–25%, 2 points for 26–50%, 3 points for 51–75% and 4 points for 75%. Besides, we classified the strength of staining was follows: non-staining—0 points, light yellow—1 point, brown yellow—2 points, and brown—3 points. Finally, we multiplied the two scores to obtain the final score which was then grouped into 4 grades: negative (0 points), weak positive (1–4 points), positive (5–8 points) and strongly positive (9–12 points). Assessment of immunohistochemical staining results was separately performed by two individuals.
Immunofluorescence
Circular glass slices were first placed in six-well plates, cell suspensions added to them, followed by a 3-h culturing in an incubator under 5% CO2 and at 37 °C to make cells climbing pieces. Cells were fixed in 4% paraformaldehyde and permeabilized with Triton X-100 PBS at room temperature for 20 minutes, then the slides washed 3 times with PBS. Absorbent paper was blotted in PBS, then 1% BSA added dropwise on the slides at room temperature for 30 min. The blocking solution was removed, then a sufficient amount of the diluted primary antibody (anti-Kiaa0101, 1:50; anti-Snail1, 1:200; anti-p-p38, 1:100) added. The contents were placed in a wet box followed by overnight incubation at 4 °C. The following day, slides were washed 3 times with PBST, diluted secondary antibody added, then incubated in a wet box at 37 °C for 1 hour in darkness. DAPI (ANT046, Antgene, 10 μg/mL) was added in the dark for 5 min, the slides washed 4 times with PBST for 5 min to remove extra DAPI, then observed under fluorescence microscope to acquire images.
Cells and cell culture
U251 and U87 cell lines were purchased from the Cell Bank Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China), and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Gibco, Invitrogen, Carlsbad, CA, USA). The cultures were incubated at 37 °C under a humidified atmosphere of 5% CO2.
Bioinformatics analysis
To clarify the expression of Kiaa0101 in glioma, we used the Gliovis database (http://gliovis.bioinfo.cnio.es/), a database based on The Cancer Genome Atlas (TCGA, http://www.tcga.org/). In this database we analyzed the differential expression of Kiaa0101 in human glioma and its impact on survival time. GEO datasets (https://www.ncbi.nlm.nih.gov/geo/) including GSE4290 and GRAVENDEEL were also used to analyze the expression of Kiaa0101 in glioma. GSE4290 was a glioma dataset which contained 23 normal brain samples and 157 glioma samples. China glioma genome atlas (CGGA, http://www.cgga.org.cn/) is the largest glioma database in China and the database contains hundreds of samples. We used this dataset to verify the oncogenic role of Kiaa0101 in glioma.
Wound healing
Wound healing was used to investigate the effect of Kiaa0101 on glioma migration. Glioma cells transfected with shRNA were seeded in 6-well plate. When cells were cultured to achieve 90% confluency, medium was aspirated and plates were washed PBS. Then we drew a scratch with a yellow tip (200 µL) vertically according to the horizontal line drew on the back of the 6-well plate. After washing with PBS for 3 times, glioma cells were cultured in FBS-free DMEM medium. The photographs were taken with an inverted microscope at 12, 24, 36, and 48 hours after scratching.
Transwell assay
Transwell chambers and Matrigel were purchased from Corning (Corning, USA) and BD Biosciences (BD, USA), respectively. Certain number of cells were cultured in FBS-free DMEM medium in the upper chamber pre-packaged with Matrigel. A 600 µL of DMEM medium with 10% FBS was placed in lower chamber. The cells were cultured for 24 hours in an incubator. Cells were fixed with 4% paraformaldehyde and stained with 1% crystal violet. Cells were finally photographed under an inverted microscope. We randomly selected 6 fields to count the number of invading cells in each set of experiments.
RNA isolation and RT-PCR
Total RNA from GBM tissues, U251 and U87 cell lines was extracted using the TRIzol regent (Invitrogen). The RNA was converted to cDNA using the PrimeScript RT reagent kit with gDNA Eraser (Takara, Tokyo, Japan). The cDNA was subjected to real-time PCR using the SYBR Green II Mixture (TaKaRa) according to the manufacturer’s protocol, targeting the KIAA0101 gene and using the following primer pair (forward 5'-TCCTGAAGAGGCAGGAAGCAGT-3'; reverse 5'-TTGTGTGATCAGGTTGCAAAGGA-3'). GAPDH (forward 5'-ACAACTTTGGTATCGTGGAAGG-3'; reverse 5'-GCCATCACG CCACAGTTTC-3') was also included as an internal amplification control.
WB
U251 and U87 cells were lysed in a modified RIPA buffer on ice for about 30 min, then centrifuged at 12,000 rpm for 15 min. Proteins from glioma tissues were extracted after grinding them under liquid nitrogen. Protein concentration was determined by BCA protein assay. The cell lysate was mixed with loading buffer after heated at 100 °C for 5 min. Equal protein amounts were loaded on a 10–15% SDS-PAGE, then transferred to a PVDF membrane. The membrane was then blocked in 5% non-fat milk for 1 hour, followed by overnight incubation with primary antibodies at 4 °C. The primary antibodies used in this study are shown in Table 1. The contents were then incubated with secondary antibody (Lincoln, NE, USA; 1:10,000) in darkness for 1 h. The experiment was performed in triplicate.

Full table
Construction and transfection with shRNA and plasmid
Oligonucleotides encoding shRNA targeted Kiaa0101 and a scrambled shRNA were purchased from Genechem Co., Ltd (Shanghai, China). The target sequence against human Kiaa0101 was 5'-GGTCCACGAATCACTATCCAC-3' and scramble shRNA was cloned into the pLKO.1 lentiviral vector. U251 and U87 cells were transfected with Lentivirus, followed by construction of Kiaa0101 overexpression plasmid and a blank vector. Cells were transfected with 2 µg plasmid using Lipofectamine 3000 and P3000 reagents (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
In vivo experiments
Thirteen 5-week-old Balb/c nude male mice, purchased from Shulaibao (Wuhan, China) Biotechnology Co., Ltd, were used in the assays, due to their immunodeficiency to prevent rejection of xenogeneic glioma cells. Animal feeding and experimental operations were performed under a project license (No. 20191005: the license number) granted by institutional ethics board of Laboratory Animal Welfare & Ethics Committee. All animals were raised in a SPF level environment, then stably transfected with U87 cells grown in a logarithmic phase and resuspended with PBS. Briefly, 100 µL of cell resuspension (5×106 cells) were subcutaneously injected into the armpits of the 5-week-old Balb/c nude mice. The mice (weighing 17–20 g) were randomly divided into two groups and their condition monitored daily. The size of xenografted tumors were calculated, as longest diameter × shortest diameter2/2. The animals were sacrificed on the 31st day after transplantation, and the tumors weighed.
Statistical analysis
Data were presented as means ± standard deviations (SD) of the means from at least three experiments. Differences between groups were determined using a student’s t-test, One-way analysis of variance (ANOVA) used for comparisons among three or more groups, whereas the student-Newman-Keuls (SNK) method was adopted for post-analysis. Patients were divided into high and low groups, according to the 50% cutoff point of Kiaa0101 expression in public datasets, and the Kaplan-Meier method used to analyze their survival times. Statistical analyses were performed in SPSS 21.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA) software, with P values less than 0.05 considered statistically significant.
Results
Kiaa0101 is overexpressed in glioma tissues
We analyzed Kiaa0101 expression patterns in different brain tissues using datasets from TCGA, GSE4290 and Rembrandt, to ascertain its role in glioma progression. Results showed that Kiaa0101 was overexpressed in glioma relative normal brain tissues (Figure 1A,B,C). Furthermore, RT-PCR and WB confirmed the above patterns, with higher levels observed in glioma than in non-tumor brain tissues, both at mRNA and proteins levels in our validation cohort (Figure 1D,E,F,G).

Kiaa0101 expression is associated with malignancy of glioma
According to the World Health Organization (WHO), gliomas are divided into four grades. The degree of malignancy increases with increase in tumor grade. Screening the aforementioned public databases revealed that Kiaa0101 increased with increase in tumor grade (Figure 2A,B,C,D). Additionally, IDH1, one of homodimeric NADP+-dependent enzymes, was an important marker in diagnosing glioma and predicting patient prognosis. Previous studies have shown that presence of an isocitrate dehydrogenase (IDH1/2) mutation in gliomas is indicative of favorable outcomes, relative to wild type gliomas (18). In fact, our results corroborated these finding, with significantly higher Kiaa0101 expression found in IDH1 wildtype gliomas relative to IDH1 mutant gliomas across three public datasets (Figure 2E,F,G). Experimental results from our validation cohort showed that high grade glioma (HGG) expressed a high constitutive level of Kiaa0101, at both mRNA and protein levels (Figure 2H,I,J, P<0.05). Correlation analysis showed revealed no association between Kiaa0101 expression with age (P=0.340), gender (P=0.275), tumor location (P=0.685) and preoperative KPS (P=0.545) (Table 2).


Full table
Next, we used Kaplan-Meier method to investigate the prognostic effect of Kiaa0101 on glioma in datasets across the aforementioned databases. Results revealed a significant association between high Kiaa0101 expression with worse overall survival across the datasets (Figure 2K,L,M,N). Furthermore, Cox regression analysis revealed that Kiaa0101 was an independent risk factor for overall survival in patients with glioma (HR =1.75, 95% CI, 1.17–2.62, P=0.006) (Table 3). These results indicate that Kiaa0101 is a novel prognostic biomarker in glioma.
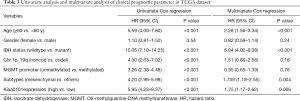
Full table
Kiaa0101 promoted migration and invasion of glioma cells in vitro
The invasive nature of tumor cells is one of the most important factors that contributes to drug resistance and tumor recurrence (19). We investigated Kiaa0101’s oncogenic role in progression of glioma using transwell and wound healing assays in U87 and U251 glioma cells. Results showed that sh-Kiaa0101 effectively downregulated Kiaa0101 expression, whereas Kiaa0101 plasmid upregulated its expression in U87 and U251 (Figure 3A). In addition, silencing Kiaa0101 in U87 and U251 significantly affected formation of the cytoskeleton by inhibiting formation of stress fibers. Moreover, Kiaa0101 knockdown resulted in reduced fluorescence intensity of phalloidin-labeled F-actin (Figure 3B), and inhibited migration and invasion of glioma cells (Figure 3C,D,E,F). On the other hand, overexpressing Kiaa0101 enhanced migration and invasion in U87 and U251 (Figure 3G,H,I,J). These results indicated that Kiaa0101 is a key molecular switch that promotes glioma migration/invasion.

Kiaa0101 induced invasion and upregulated Snail1
Snail1 is a crucial transcription factor in epithelial-mesenchymal transition, an important biological process through which malignant tumor cells are generated from the epithelium and obtain migration and invasion abilities. Our results revealed a positive correlation between Kiaa0101 with Snail1, Vimentin and mmp2 in TCGA (Figure 4A). WBs showed that silencing Kiaa0101 reduced expression of Snail1, mmp2 and Vimentin in both U87 and U251 cells (Figure 4B,C). Additionally, overexpressing Kiaa0101 in glioma cells up-regulated Snail1, mmp2 and Vimentin expression (Figure 4B,C,D). These results were validated by immunofluorescence, with results revealing a downregulation of snail1 in both the nucleus and cytoplasm following Kiaa0101 inhibition (Figure 4E). Immunohistochemistry (IHC) analysis revealed that high snail1 expression (Figure 4F,G), which was more common in gliomas that overexpressed Kiaa0101, relative to tumors with low or no expression.

Kiaa0101 enhanced invasion through Snail1
To determine whether Snail1 is the key transcription factor in Kiaa0101-mediated invasion in glioma cells, we used siRNAs to downregulate Snail1 expression in cells transfected with Kiaa0101 vector. Results showed that silencing snail1 in Kiaa0101-transfected glioma cells significantly counteracted Kiaa0101 effect on promoting migration and invasion in both U251 (Figure 5A,B) and U87 cells (Figure 5C,D). WBs showed that si-snail1 could also downregulate snail1, mmp2 and vimentin expression in Kiaa0101 stably expression glioma cells (Figure 5E,F). Meanwhile, immunofluorescence (IF) staining showed the same results (Figure 5G). Taken together, these results indicated that snail1 plays a crucial role in mediating Kiaa0101-promoting invasion.

Kiaa0101 regulates migration and invasion of glioma cells via the p38 MAPK signaling pathway
The MAPK pathway plays a key role in regulating migration and invasion. In addition, previous studies have implicated p38 in regulating invasion in glioma cells (20-22). In this study, investigated the effect of Kiaa0101 on p38 MAPK. Summarily, knocking down Kiaa0101 in glioma cells significantly downregulated p-p38 and snail1 expression, but had no effect on p38 expression (Figure 6A). Conversely, upregulating Kiaa0101 upregulated p-p38 and snail1 in U87 and U251 (Figure 6A). To further validate our findings, we preformed IF staining and found that downregulating Kiaa0101 expression decreased nuclear accumulation of p-p38 (Figure 6B).

To investigate the role of p38 MAPK signaling in mediating Kiaa0101 enhanced invasion, we used a p38 MAPK signaling inhibitor, BIRB796. BIRB796’s effect on p38 MAPK signaling was evaluated in our previous study (18). In the present study, Kiaa0101-transfected glioma cells treated with BIRB796 dramatically downregulated snail1 expression thereby inhibiting invasion of U87 and U251 (Figure 6C,D,E,F). These results indicated that Kiaa0101 mediated p38 MAPK signaling activation might be a potential mechanism through which Kiaa0101 promotes glioma malignancy. Moreover, we investigated the relationship between Kiaa0101 and p-p38 by following their detection in glioma tissue. WB and IHC results revealed a positive correlation between Kiaa0101 expression with p-p38 levels (Figure 6G,H,I,J).
Kiaa0101 slicing suppressed glioma malignancy in vivo
To further ascertain the role of Kiaa0101 in promoting glioma malignancy, we established subcutaneous xenograft models, with 6 and 7 mice categorized into NC and sh-Kiaa0101 groups, respectively. Nude mice in both groups developed subcutaneous solid tumors after 5 days, and were all sacrificed 29 days after transplantation (Figure 7A). Our results showed that tumors in the sh-Kiaa0101 group had a significantly lower growth rate than those in the NC group (Figure 7B). Similarly, tumors in the sh-Kiaa0101 group had significantly lower weights (0.36±0.20) relative to those in the NC group (1.24±0.37) (P=0.002, Figure 7C). Additionally, snail1, mmp2, vimentin and p-p38 were all downregulated in mice in the sh-Kiaa0101 group (Figure 7D).

Discussion
According to the WHO classification, gliomas can be divided into four grades. Glioblastoma (WHO IV) which accounts for 56.6% of all gliomas has fetal clinical prognosis (2). Prognosis of patients with low grade gliomas (LGGs) is better than that of those with glioblastoma, while the majority of LGG patients eventually progress to high grade gliomas (HGG) (23,24). Significant efforts have been made to identify biomarkers closely associated with glioma progression and clinical prognosis. Such molecular markers are critical to understanding the classification, diagnosing and treatments of gliomas. For example, Butz et al. (25) reported that significantly higher Kiaa0101 expression in renal clear cell carcinoma (ccRCC) compared with normal tissues, using datasets from public databases. Furthermore, higher Kiaa0101expression was associated with lower disease-free survival, suggesting that is may be an independent prognostic marker for both ccRCC and pancreatic cancer (26,27). However, only a handful of studies have focused on Kiaa0101’s oncogenic role in glioma. For example, Derrick et al. reported that Kiaa0101 was an important DNA damage response gene, following its upregulation in GBM relative to grade glioma, using the TCGA dataset and tissue microarrays. Subsequently survival analysis, solely based on Rembrandt, revealed a negative correlation between Kiaa0101 with clinical outcomes (7). Results of the present study indicate that Kiaa0101 was not only upregulated in higher grade glioma, but was also associated with IDH status. Additionally, the high Kiaa0101 expression in glioma was significantly correlated with worse overall survival across all four public datasets. Overall, these results indicated that high Kiaa0101 expression was as an independent risk factor for poor prognosis in glioma.
Previous studies have described the oncogenic role of Kiaa0101 in several solid tumors, indicating that it is a regulator of cell proliferation both in vivo and in vitro (6,26). For instance, Kiaa0101 was found to be mainly localized in the nucleus, where it plays a crucial role in regulating DNA replication and cell survival (28). Moreover, Kiaa0101 interacted with PCNA, subsequently promoted glioma stem cell self-renewal and radio-resistance (11). Initially, the infiltrative nature of glioma cells is manifested as a malignant tumor, which leads to resistance to chemotherapy and early recurrence (29). However, Kiaa0101’s role in promoting invasion of glioma has seldom been investigated. Results from the present study showed that Kiaa0101 promoted migration and invasion of glioma cells by regulating expression of snail1. Bioinformatics analysis further revealed a close association between Kia0101 expression in gliomas and snail1. The transcriptional activity of Snail1 mainly depends on its distribution in cells, whereas post-translational phosphorylation, ubiquitination, and lysine oxidation all influence Snail1 protein stability, subcellular localization, and activity. In the present study, it was evident that snail1 expression and subcellular location were significantly altered in sh-Kiaa0101 transfected cells, further confirming that snail1 mediates Kiaa0101-promoting invasion. However, the underlying transcriptional or post-transcriptional mechanisms regulating snail1 remain unknown, and require further investigation. Our findings were consistent with previous studies by Jain et al. (30), who revealed that siRNA-mediated Kiaa0101 knockdown significantly decreased invasion of NCI-H295R in vitro. Similarly, Kiaa0101 knockdown was found to significantly inhibit migration and invasion of SKOV3 and COV644 cells by inducing expression of E-cadherin and N-cadherin (31). Overall, these results affirm Kiaa0101’s role as a crucial promoter for invasion of glioma, indicating that it is a potential target in treatment of glioma.
Kiaa0101’s variant 1 was reported to inhibit activation of p53 by directly binding, thereby arresting doxorubicin-induced apoptosis (32). Moreover, variant 2 was shown to compete with variant 1 in binding p53, which led to reduced proliferation and invasion via elevated p53 activity (33). In the present study, Kiaa0101 was an important regulator of phosphorylation of the p38MAPK signaling pathway, a major regulator of biological processes, such as inflammation, tumorigenesis and stemness maintenance (34,35). Previous studies have shown that inhibiting p38 MAPK signaling reduces invasion of glioma cells. Rodríguez et al. (36) found that p38 MAPK signaling was essential for IL-1β-induced RUNX1 expression, which subsequently enhanced invasion of glioma cells. In addition, p38 translocation in the cytoplasm and nucleus is an important process that activates the p38 MAPK signaling pathway. Consequently, phosphorylation of p38 in the nucleus causes p-p38 to activate its substrates, such as MK2, MK3 and MK5 (12). In addition, Wang et al. (37) found that Kiaa0101 enhanced proliferation of Bel7402 cells, and siRNA-mediated knock down markedly inhibited MEK/ERK signaling; a crucial downstream component of MAPK cascades. Our findings indicated that Kiaa0101 might have activated p38 signaling, which subsequently increased snail1expression, thereby enhancing migration and invasion of glioma cells. Notably, results from IF staining also revealed significantly lower p-p38 accumulation in the nucleus following Kiaa0101 inhibition in glioma cells. These findings suggested that Kiaa0101 is a regulator of p-p38, hence could be a potential therapeutic target. Kiaa0101 may affect intracellular level of p-p38 by post-transcriptional modification of p38. Since p38 lacks both nuclear localization sequence (NLS) and a nuclear export sequence (NES), we hypothesized that Kiaa0101 might act as a chaperone protein that enables nuclear translocation of p38. However, it is unclear whether Kiaa0101 directly binds to p38 to affect its phosphorylation or regulate its nuclear exportation. Future researches are therefore expected to evaluate the interaction between Kiaa0101 and p38.
In conclusion, our findings indicate that Kiaa0101 is upregulated in glioma, especially in HGGs. Apart from correlating with worse overall survival, higher Kiaa0101 expression is an independent risk factor for prognosis of glioma patients. Furthermore, knocking down Kiaa0101 reduced the migration and invasion of glioma cells by regulating the p38 MAPK signaling pathway. Taken together, these results demonstrated that Kiaa0101 is a novel prognostic biomarker, and inhibiting it might be an effective strategy for treating glioma.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 81572489).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-3219
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-3219
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-3219
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3219). The authors have no interests to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All patients signed informed consents and this study was approved by the Institutional Ethics Committee of the Faculty of Medicine at Renmin Hospital of Wuhan University [approval number: 2012LKSZ (010) H]. Animal feeding and experimental operations were performed under a project license (No. 20191005: the license number) granted by institutional ethics board of Laboratory Animal Welfare & Ethics Committee.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Davis FG, Smith TR, Gittleman HR, et al. Glioblastoma incidence rate trends in Canada and the United States compared with England, 1995-2015. Neuro Oncol 2020;22:301-2. [Crossref] [PubMed]
- Ostrom QT, Cote DJ, Ascha M, et al. Adult Glioma Incidence and Survival by Race or Ethnicity in the United States From 2000 to 2014. JAMA Oncol 2018;4:1254-62. [Crossref] [PubMed]
- Lefranc F, Le Rhun E, Kiss R, et al. Glioblastoma quo vadis: Will migration and invasiveness reemerge as therapeutic targets? Cancer Treat Rev 2018;68:145-54. [Crossref] [PubMed]
- Zhang X, Zhang W, Mao XG, et al. Targeting role of glioma stem cells for glioblastoma multiforme. Curr Med Chem 2013;20:1974-84. [Crossref] [PubMed]
- Yu P, Huang B, Shen M, et al. p15(PAF), a novel PCNA associated factor with increased expression in tumor tissues. Oncogene 2001;20:484-9. [Crossref] [PubMed]
- Lv W, Su B, Li Y, et al. KIAA0101 inhibition suppresses cell proliferation and cell cycle progression by promoting the interaction between p53 and Sp1 in breast cancer. Biochem Biophys Res Commun 2018;503:600-6. [Crossref] [PubMed]
- Yuan RH, Jeng YM, Pan HW, et al. Overexpression of KIAA0101 predicts high stage, early tumor recurrence, and poor prognosis of hepatocellular carcinoma. Clin Cancer Res 2007;13:5368-76. [Crossref] [PubMed]
- Jin C, Liu Z, Li Y, et al. PCNA-associated factor P15(PAF), targeted by FOXM1, predicts poor prognosis in high-grade serous ovarian cancer patients. Int J Cancer 2018;13:5368-76.
- Abdelgawad IA, Radwan NH, Hassanein HR. KIAA0101 mRNA expression in the peripheral blood of hepatocellular carcinoma patients: Association with some clinicopathological features. Clin Biochem 2016;49:787-91. [Crossref] [PubMed]
- Su X, Zhang T, Cheng P, et al. KIAA0101 mRNA overexpression in peripheral blood mononuclear cells acts as predictive marker for hepatic cancer. Tumour Biol 2014;35:2681-6. [Crossref] [PubMed]
- Ong DS, Hu B, Ho YW, et al. PAF promotes stemness and radioresistance of glioma stem cells. Proc Natl Acad Sci U S A 2017;114:E9086-E9095. [Crossref] [PubMed]
- Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev 2011;75:50-83. [Crossref] [PubMed]
- Bedard PL, Tabernero J, Janku J, et al. A phase Ib dose-escalation study of the oral pan-PI3K inhibitor buparlisib (BKM120) in combination with the oral MEK1/2 inhibitor trametinib (GSK1120212) in patients with selected advanced solid tumors. Clin Cancer Res 2015;21:730-8. [Crossref] [PubMed]
- Ho AL, Grewal RK, Leboeuf R, et al. Selumetinib-enhanced radioiodine uptake in advanced thyroid cancer. N Engl J Med 2013;368:623-32. [Crossref] [PubMed]
- Tatevossian RG, Lawson AR, Forshew T, et al. MAPK pathway activation and the origins of pediatric low-grade astrocytomas. J Cell Physiol 2010;222:509-14. [PubMed]
- Mason WP, Belanger K, Nicholas G, et al. A phase II study of the Ras-MAPK signaling pathway inhibitor TLN-4601 in patients with glioblastoma at first progression. J Neurooncol 2012;107:343-9. [Crossref] [PubMed]
- Schiff D, Jaeckle KA, Anderson SK, et al. Phase 1/2 trial of temsirolimus and sorafenib in the treatment of patients with recurrent glioblastoma: North Central Cancer Treatment Group Study/Alliance N0572. Cancer 2018;124:1455-63. [Crossref] [PubMed]
- Huse JT, Aldape KD. The evolving role of molecular markers in the diagnosis and management of diffuse glioma. Clin Cancer Res 2014;20:5601-11. [Crossref] [PubMed]
- Liu CA, Chang CY, Hsueh KW, et al. Migration/Invasion of Malignant Gliomas and Implications for Therapeutic Treatment. Int J Mol Sci 2018;19:1115. [Crossref] [PubMed]
- Pandey V, Bhaskara VK, Babu PP. Implications of mitogen-activated protein kinase signaling in glioma. J Neurosci Res 2016;94:114-27. [Crossref] [PubMed]
- Yuan F, Liu B, Xu Y, et al. TIPE3 is a regulator of cell apoptosis in glioblastoma. Cancer Lett 2019;446:1-14. [Crossref] [PubMed]
- Liao J, Tao X, Ding Q, et al. SSRP1 silencing inhibits the proliferation and malignancy of human glioma cells via the MAPK signaling pathway. Oncol Rep 2017;38:2667-76. [Crossref] [PubMed]
- Claus EB, Walsh KM, Wiencke JK, et al. Survival and low-grade glioma: the emergence of genetic information. Neurosurg Focus 2015;38:E6. [Crossref] [PubMed]
- Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010;17:98-110. [Crossref] [PubMed]
- Butz H, Szabo PM, Nofech-Mozes R, et al. Integrative bioinformatics analysis reveals new prognostic biomarkers of clear cell renal cell carcinoma. Clin Chem 2014;60:1314-26. [Crossref] [PubMed]
- Fan S, Liu X, Tie L, et al. KIAA0101 is associated with human renal cell carcinoma proliferation and migration induced by erythropoietin. Oncotarget 2016;7:13520-37. [Crossref] [PubMed]
- Hosokawa M, Takehara A, Matsuda K, et al. Oncogenic role of KIAA0101 interacting with proliferating cell nuclear antigen in pancreatic cancer. Cancer Res 2007;67:2568-76. [Crossref] [PubMed]
- Xie C, Yao M, Dong Q. Proliferating cell unclear antigen-associated factor (PAF15): a novel oncogene. Int J Biochem Cell Biol 2014;50:127-31. [Crossref] [PubMed]
- Schiffer D, Annovazzi L, Casalone C, et al. Glioblastoma: Microenvironment and Niche Concept. Cancers (Basel) 2018;11:5. [Crossref] [PubMed]
- Jain M, Zhang L, Patterson EE, et al. KIAA0101 is overexpressed, and promotes growth and invasion in adrenal cancer. PLoS One 2011;6:e26866. [Crossref] [PubMed]
- Chen H, Xia B, Liu T, et al. KIAA0101, a target gene of miR-429, enhances migration and chemoresistance of epithelial ovarian cancer cells. Cancer Cell Int 2016;16:74. [Crossref] [PubMed]
- Liu L, Chen X, Xie S, et al. Variant 1 of KIAA0101, overexpressed in hepatocellular carcinoma, prevents doxorubicin-induced apoptosis by inhibiting p53 activation. Hepatology 2012;56:1760-9. [Crossref] [PubMed]
- Liu L, Liu Y, Chen X, et al. Variant 2 of KIAA0101, antagonizing its oncogenic variant 1, might be a potential therapeutic strategy in hepatocellular carcinoma. Oncotarget 2017;8:43990-4003. [Crossref] [PubMed]
- Corre I, Paris F, Huot J. The p38 pathway, a major pleiotropic cascade that transduces stress and metastatic signals in endothelial cells. Oncotarget 2017;8:55684-714. [Crossref] [PubMed]
- Cuadrado A, Nebreda AR. Mechanisms and functions of p38 MAPK signalling. Biochem J 2010;429:403-17. [Crossref] [PubMed]
- Sangpairoj K, Vivithanaporn P, Apisawetakan S, et al. RUNX1 Regulates Migration, Invasion, and Angiogenesis via p38 MAPK Pathway in Human Glioblastoma. Cell Mol Neurobiol 2017;37:1243-55. [Crossref] [PubMed]
- Wang Q, Wang Y, Li Y, et al. NS5ATP9 contributes to inhibition of cell proliferation by hepatitis C virus (HCV) nonstructural protein 5A (NS5A) via MEK/extracellular signal regulated kinase (ERK) pathway. Int J Mol Sci 2013;14:10539-51. [Crossref] [PubMed]


