
Establishment of multiplex allele-specific blocker PCR for enrichment and detection of 4 common EGFR mutations in non-small cell lung cancer
Introduction
Lung cancer has the highest incidence and mortality among all cancers globally (1). Around 85% of cases are non-small cell lung cancer (NSCLC) (2). For patients with lung cancer, surgery is the most effective therapy, but unfortunately nearly 70% of patients have locally advanced or metastasized cancer when diagnosed (3), so they have to rely on other therapies. However, lung cancer shows genetic susceptibility (4), which provides opportunity for personalized therapy and precision medicine.
New findings of changes in the main signal and regulation pathway (overexpression or sequence variation) during carcinogenesis and development provide opportunities for targeted therapy (3,5), which, compared with traditional chemotherapy, has better specificity, lower cytotoxicity and higher efficacy (5,6). In 40–80% of NSCLC patients, epidermal growth factor receptor (EGFR) is overexpressed (7), so they might benefit from EGFR inhibitors incorporating tyrosine kinase inhibitors (TKI) (6) and monoclonal antibodies (8). In almost 70% of cases, patients’ overall survival is prolonged with EGFR-TKI (9), but many patients show resistance to certain drugs, which is a serious challenge for targeted therapy (10). Most EGFR mutations are located in exons 18–21 of the coding region of the TK domain (11,12), and they are related to a patient’s sensitivity to TKI (12). The most common drug-sensitive variations are exon 19 deletion (19del) and exon 21 L858R, which comprise 45% and 40–45% respectively of EGFR mutations in NSCLC (12). Currently, gefitinib and erlotinib are approved by the United States Food and Drug Administration (FDA) as first-line therapy for advanced NSCLC patients carrying the EGFR 19del or L858R mutation (3,13). However, with progression of disease, the T790M mutation in exon 20, which is associated with acquired resistance to 1st- and 2nd-generation EGFR TKI, can be detected in 50% of patients (9,12,13). A 3rd-generation TKI, osimertinib, is also approved by FDA because of its strong effectiveness in patients with the T790M mutation (14,15). Thus, precise diagnosis of these mutations can guide clinical decision-making regarding therapy.
Tissue biopsy is the golden standard for obtaining tumor genetic information, but invasive sampling, DNA-damaging operation and tumor heterogeneity restrain its utilization (16,17). Liquid biopsy overcomes these shortcomings by detecting circulating tumor cell (CTC), cell-free DNA (cfDNA) and other tumor biomarkers from blood or body fluids. Circulating tumor DNA (ctDNA) is a type of cfDNA derived from cancer cells (18), so it contains many tumor-related biomarkers such as methylation changes (19-22), single-nucleotide polymorphisms (23-27), copy number variations (20,28-30) and chromosomal rearrangement (31,32). Nevertheless, ctDNA is highly fragmented and has a massive wild-type background, which poses a big challenge to enrichment and detection technology. Common minor-allele enriching strategies include amplification refractory mutation system (ARMS) (33), blocker PCR (34) or clamping PCR (35-37), and co-amplification at lower denaturation temperature-PCR (COLD-PCR) (38-40). However, all these strategies are low in throughput, which increases the difficulty of operation and heightens the risk of contamination while decreasing the detection efficiency. There are some improved or combined strategies such as SuperSelective primer (41), improved and complete enrichment COLD-PCR (ice COLD-PCR) (42), and nuclease-assisted minor-allele enrichment with probe-overlap (NaME-PrO) (43), but they have not been widely used yet. For ctDNA detection, the mainstream technologies are digital PCR (dPCR) (44), beads, emulsion, amplification and magnetics (BEAMing) (45,46), sequencing (47,48) and quantitative PCR (qPCR) (49). dPCR, BEAMing and sequencing rely on specialized apparatus as well as complicated operation and analysis procedures. In addition, their high cost restricts their clinical application. qPCR is sensitive enough [0.1–1% mutant allele fraction (MAF)] (50) for ctDNA detection, while being fast and inexpensive, so it is widely applied in clinical settings (49).
We established a novel method called multiplex allele-specific blocker PCR (MAB PCR), which can accomplish multiplex enrichment and detection of mutations from ctDNA. The target mutations included the most common 19del mutations (c.2235_2249del-15 and c.2236_2250del-15), and the T790M and L858R mutations. This method provides a novel solution for quick and accurate detection of these 4 target mutations from clinical samples, especially ctDNA. It has huge potential in guiding precision medicine for NSCLC patients. We present the following article in accordance with the STARD reporting checklist (available at http://dx.doi.org/10.21037/atm-20-6754).
Methods
Sample collection and preparation
Two types of samples were included in this research: frozen tissue samples and plasma samples collected from Shanghai Pulmonary Hospital. Sample donors gave written informed consent before enrollment. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics committee of Shanghai Pulmonary Hospital (No.: K18-040-1).
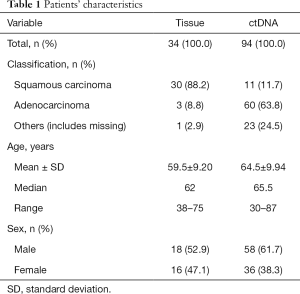
Frozen tissue samples (n=34) were collected between November 2016 and January 2018 (Table 1) from patients’ tumor tissues, and DNA was extracted by QIAamp DNA Mini Kit (Qiagen, Germany). The concentration and quality of DNA was measured by Spectrophotometer ND-2000 (Thermo Fisher Scientific, USA). A 10-µL DNA dilution of every sample was sequenced (Jie Li Biology, Shanghai, China) and the remaining DNA dilution was stored at –20 °C.
Full table
The 94 plasma samples were obtained between March 2017 and January 2018 (Table 1) by the following process: 5 mL peripheral blood sample was collected into ethylenediaminetetraacetic acid (EDTA) tubes, then centrifuged at 2,000 g for 10 min at 4 °C and the plasma was transferred into a new centrifuge tube. The ctDNA was extracted by cfDNA Isolation Kit (Shanghai Realgen Biotech, China) and its concentration was measured by quantifying β-actin using qPCR. Isolated ctDNA was stored at –20 °C.
Design of the MAB PCR
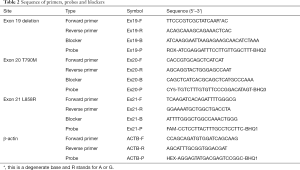
Because ctDNA is of low concentration and highly fragmented, to improve the performance of enrichment, MAB PCR combined ARMS and blocker PCR. For each mutation site, an allele-specific primer (AS-primer) that matched the mutant-type sequence and a blocker that matched the wild-type sequence were designed (for the two deletions on exon 19, a degenerate base was adopted on the primer so it could amplify both deletions on exon 19). The blockers covered the mutation sites and their 3’ends were sealed so that they could not be extended. At a certain melting temperature (at which half of the DNA strands are in the random coil or single-stranded state), the AS-primer preferred the mutant-type template while the blocker preferred wild-type template, which caused discrimination in amplification and therefore the mutant-type template was enriched. Considering the length of ctDNA fragments is usually around 167 base pairs (bp), the reverse primer was specially designed so that the amplicon (range, 66–106 bp) would be within 1 fragment. To promote the throughput, fluorescent probes were applied to accomplish multiplex detection in 1 reaction tube. The reference gene was β-actin. Primers and blockers were designed by Primer Premier 5 (PREMIER Biosoft International). All oligonucleotides were synthesized by Shanghai Generay Biotech. The sequence of all primers, blockers and probes are showed in Table 2.
Full table
Optimization and evaluation of MAB PCR
The assay was performed in a MA-6000 Real-Time PCR Machine (Molarray, China). In order to optimize the performance of MAB PCR, plasmids containing every mutant-type target sequence were constructed. Genomic DNA (extracted from a 293T cell line) was applied as the wild-type template. As there were four sets of primers and probes, we had to optimize every singleplex reaction first and group up the 4 singleplex reactions in 1 tube for final optimization.
After optimizing the PCR ingredient and thermal cycling conditions, we used serially diluted plasmids (range, 100–106 copies) to test the limit of detection (LOD). To determine the sensitivity of this assay, serially diluted mutant-type plasmids (range, 100–105 copies) were 1:1 mixed with 330 ng human genomic DNA (293T cell line) to generate mixed templates containing 0%, 0.001%, 0.01%, 0.1%, 1%, 10%, and 50% mutant DNA respectively. Each sample was analyzed in triplicate. After the determination of sensitivity, the cycle number would be used to calculate the threshold and when a clinical sample’s Cq (quantification cycle) was larger than the threshold, it would be called wild-type.
Comparison of singleplex and multiplex assays
According to the sequencing results, 13 mutant- and 7 wild-type tissue samples were chosen for comparison of the performance of multiplex and singleplex assays at each site. The 13 mutant-type samples included 6 samples with exon 19 deletion, 2 samples with exon 20 T790M and 6 samples with exon 21 L858R, within which a sample contained the exon 19 deletion (c.2235_2249del-15) and exon 20 T790M simultaneously. The wild-type samples were randomly chosen from all wild-type samples. These 20 samples were amplified respectively by singleplex and multiplex assays concurrently and the data were statistically analyzed by Pearson correlation analysis and Bland-Altman method to test the consistency of performance.
Clinical validation and statistical analysis
We used MAB PCR to screen and profile clinical samples (both tissue and ctDNA). The results for the tissue samples were validated by sequencing, whereas the results for ctDNA screening were compared with the sequencing data from the patient’s corresponding tissue sample. The accuracy was calculated to assess the performance of this assay.
Results
Quality control of samples
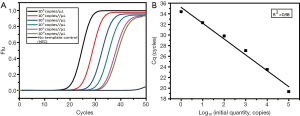
The concentration of tissue sample DNA ranged from 20.87 to 779.89 ng/µL, and all were diluted to 20 ng/µL for screening. By comparing the Cq of samples and the standard curve (Figure 1), the copy number of ctDNA could be quantified. The concentration range of ctDNA was 104–103 copies/µL.
Sensitivity of the assay
First, the singleplex assays for each mutant site were built and optimized. The LOD of these three assays were tested respectively by serially diluted plasmids (Figure 2). For each site, we plotted the Cq against plasmid concentration to give the standard curve, from which the linearity and amplification efficiency were calculated. Every singleplex assay showed good linearity and high efficiency, which proved they could be assembled into one tube.

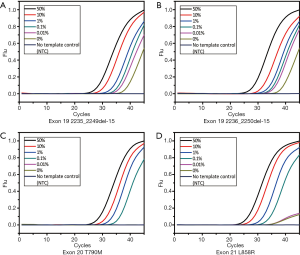
The multiplex assay consisted of the three previously mentioned singleplex assays as well as the β-actin primers and probe set. The final volume of this assay was 25 µL. The working solution contained 1× PCR buffer [670 mM Tris-HCl (pH 8.0), 160 mM (NH4)2SO4 and 0.1% Tween-20 (w/v)], 4.5 mM MgSO4, 0.25 mM dNTPs, 2.0 U of hot-start Taq polymerase (TaKaRa, Dalian, China), 0.12–0.14 µM primers and probes, 0.04–0.16 µM blockers and 1 µL of DNA sample. The thermal conditions of PCR were as follow: pre-denaturation at 95 °C for 10 min, a 2-step amplification with 45 cycles of denaturation at 95 °C for 15 s, and annealing and extension at 60 °C for 60 s. The fluorescence from each channel (FAM, ROX, HEX and CY5) was acquired at the end of the annealing and extension step. This assay was tested by serially diluted mutant-type plasmids of each site to determine the LOD and amplification capability (Figure 3A). Similar to the singleplex assays, the standard curve was plotted to calculate the linearity and amplification efficiency (Figure 3B). The MAB PCR assay retained the high linearity and efficiency from the singleplex assays and showed great sensitivity.
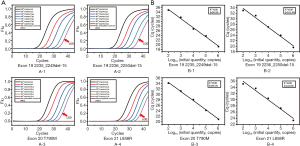
The statistical analysis of performance is shown in Table 3. For each site, amplification of the wild-type templates was suppressed and the Cq of all serial dilutions was larger than 102 copies/µL of the respective mutant-type template, although 102 copies/µL of every mutant-type template could be detected. Thus, the LOD of each site was 102 copies/µL. The R2 of each standard curve was 0.99 and the efficiency ranged from 89.5%±2.52% to 110.6%±1.85%, which proved that the amplification of each site was roughly balanced. The coefficient of variation (CV) of triplicate testing at each site was less than 5% and hence the assay was robust.

Full table
The sensitivity test helped us understand the tolerance capability of the MAB PCR assay and determine the threshold of the mutant-type allele (Figure 4). At each mutant site, template containing 0.1% mutant-type allele could be distinguished from the wild-type, so the sensitivity of this assay was 0.1% MAF. For each site, the Cq of 0.1% MAF in triplicate was calculated by (
Comparison of singleplex and multiplex assays
We collected the Cq of singleplex and multiplex assays respectively and plotted the scatter diagram using SPSS Statistics 17.0 (SPSS Inc.) (Figure 5). The Pearson correlation coefficient (r) was used to describe the consistency of singleplex and multiplex assays at each site. For each mutation site, r was higher than 0.85 (P>0.05), which suggested that the singleplex and multiplex assays were consistent.
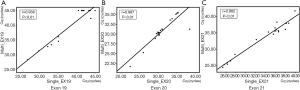
To further investigate the consistency of the singleplex and multiplex assays, the Bland-Altman method was applied (Figure 6). The analysis and diagram process were conducted in MedCalc 18.5 (MedCalc Software Ltd.). As the plot shows, the mean difference of every mutation was close to zero, which revealed low deviation, while most spots were aggregated within the 95% confidence interval of consistency. In conclusion, the singleplex and multiplex assays were highly accordant.

Clinical sample screening and statistical analysis
After screening and identification, all samples were genotyped and compared with correlated sequencing data (Table 4). There were 6 exon 19 deletions, 2 exon 20 T790M and 6 exon 21 L858R mutations detected in 34 tissue samples, which exactly matched the sequencing data. For 94 ctDNA samples, 11 exon 19 deletions, 1 exon 20 T790M and 11 exon 21 L858R mutations were distinguished, which accorded with the sequencing results for primary tumor tissue and proved that ctDNA could reflect the mutation status of the tumor.

Full table
Discussion
With the development of targeted therapy, the overall survival of many patients is now extended and their suffering is relieved. However, genetic mutations (e.g., EGFR, KRAS, BRAF, etc.) can restrict the efficacy of drugs. As precision medicine becomes widespread, new diagnostic technology, especially technology aimed at non-invasive sampling such as ctDNA, has become the hot spot of research. In particular, ctDNA can reflect the genetic features of tumor cells, and is especially suitable for early diagnosis and companion diagnostics because it can be easily obtained. For patients whose tumor tissue is either unavailable or inadequate, ctDNA would be an ideal substitute for a surgical biopsy.
Recently, more technologies have filled the armamentarium of molecular diagnosis. Multiple methods such as NGS (next-generation sequencing), microarray, dPCR and qPCR are available for detecting mutations from clinical specimens. However, when it comes to liquid biopsy, qPCR has an enormous advantage among all these techniques because it is sensitive, simple and inexpensive. Our special design, the MAB PCR assay, could detect the four most common EGFR mutations simultaneously from various types of samples. β-actin was used as the control to exclude external interference. This assay showed excellent performance with high amplification efficiency and linearity at every mutation site. The LOD of this assay was 102 copies/µL and the detection sensitivity reached 0.1% MAF, enough for ctDNA detection. In clinical sample screening, the results of MAB PCR assay indicated great consistency with the sequencing results. Overall, our design reduced the expense and simplified the operation. Compared with other methods, the MAB PCR assay was fast, accurate and robust.
The highlights of MAB PCR are as follow. First, its design combines ARMS and blocker technology. With this complex overlap, the ARMS primers and blockers had to compete to bind to the templates. The primers preferentially matched the mutant-type template, while the blockers preferred the wild-type template. The different priority of matching created discrimination in amplification and led to enrichment of the mutant-type template. Second, the application of fluorescent probes increased the throughput and decreased the risk of false-positive cases. The fluorescent probes allowed us to detect multiple targets in a single reaction and to observe the whole process of amplification. Compared with fluorescent dyes such as SYBR Green, probes are more selective and easier for accomplishing multiplicity. Moreover, the fluorescent signal could be detected with the tube sealed and replaced the post-PCR procedure, which is a major source of contamination. Third, the design of MAB PCR made the operation procedure easy and simple to operate, and it was inexpensive to conduct. The design especially catered to ctDNA, so that it could be applied in companion diagnostics. Other sample types are compatible, which increases its practicability. Overall, MAB PCR has huge potential in clinical usage, and could be further developed into a diagnostic kit for guiding medical treatment.
We suggest that the threshold of the MAB PCR assay be amended regularly, especially when there is any change in equipment, reaction reagent or conditions. Incorrect thresholds might cause mistake in genotyping and therefore lead to false-positive or false-negative cases.
The number of clinical samples used in this study was limited, especially exon 20 T790M mutant-type samples due to its rareness. Moreover, the majority of samples were from late-stage patients. For future research, more samples, particularly from patients with the exon 20 T790M mutation, should be collected to further prove the stability of this assay. Meanwhile, although this assay has theoretical capability in early diagnosis, more early-stage samples should be incorporated to determine the performance of MAB PCR when used in early diagnosis.
In addition, the design of MAB PCR can be modified, especially the design of the probes, to achieve a higher throughput with more mutant sites detected at the same time. This design could be applied to other mutation sites. We are currently considering the possibility of improving the throughput of COLD-PCR by using fluorescent probe. These new designs might have dramatic potential in liquid biopsy for both cancer and non-invasive prenatal testing (NIPT).
Acknowledgments
We really appreciate the participation of all the patients.
Funding: This study was funded by National Natural Science Foundation of China (31571371, 81772657), National Key Research and Development Project (2017YFC0907501), Shanghai Committee of Science and Technology (17ZR1423500), Shanghai Science and Technology Innovation Action Plan (17JC1400902) and the Fundamental Research Funds for the Central Universities (22120180371).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-6754
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-6754
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6754). Mr. Hongyuan C and Dr. Hongyan C report that they have a patent 2019107453518 pending. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics committee of Shanghai Pulmonary Hospital (No.: K18-040-1). Sample donors gave written informed consent before enrollment.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Qin BM, Chen X, Zhu JD, et al. Identification of EGFR kinase domain mutations among lung cancer patients in China: implication for targeted cancer therapy. Cell Res 2005;15:212. [Crossref] [PubMed]
- Lin CC. Challenges of the phase I drug development in non-small cell lung cancer. Chin Clin Oncol 2019;8:25. [Crossref] [PubMed]
- Kiyohara C, Otsu A, Shirakawa T, et al. Genetic polymorphisms and lung cancer susceptibility: a review. Lung Cancer 2002;37:241-56. [Crossref] [PubMed]
- Papadopoulos N, Kinzler KW, Vogelstein B. The role of companion diagnostics in the development and use of mutation-targeted cancer therapies. Nat Biotechnol 2006;24:985-95. [Crossref] [PubMed]
- Masuda K, Fujiwara Y, Shinno Y, et al. Efficacy and safety of crizotinib in patients with ROS1 rearranged non-small cell lung cancer: a retrospective analysis. J Thorac Dis 2019;11:2965-72. [Crossref] [PubMed]
- Passaro A, de Marinis F. Looking for the high way in EGFR-positive non-small cell lung cancer through the evaluation of survival endpoints. Transl Lung Cancer Res 2019;8:S334-8. [Crossref] [PubMed]
- Reck M, Rabe KF. Precision Diagnosis and Treatment for Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:849-61. [Crossref] [PubMed]
- Zhu L, Zhang S, Xun Y, et al. Comparison of the Amplification Refractory Mutation System, Super Amplification Refractory Mutation System, and Droplet Digital PCR for T790 M Mutation Detection in Non-small Cell Lung Cancer after Failure of Tyrosine Kinase Inhibitor Treatment. Pathol Oncol Res 2018;24:843-51. [Crossref] [PubMed]
- Tan CS, Gilligan D, Pacey S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol 2015;16:e447-59. [Crossref] [PubMed]
- Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005;352:786-92. [Crossref] [PubMed]
- Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007;7:169-81. [Crossref] [PubMed]
- Kim E, Feldman R, Wistuba II. Update on EGFR Mutational Testing and the Potential of Noninvasive Liquid Biopsy in Non-Small-cell Lung Cancer. Clin Lung Cancer 2018;19:105-14. [Crossref] [PubMed]
- Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689-99. [Crossref] [PubMed]
- Kawamura T, Kenmotsu H, Omori S, et al. Clinical Factors Predicting Detection of T790M Mutation in Rebiopsy for EGFR-Mutant Non-small-cell Lung Cancer. Clin Lung Cancer 2018;19:e247-52. [Crossref] [PubMed]
- Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Volckmar AL, Sultmann H, Riediger A, et al. A field guide for cancer diagnostics using cell-free DNA: From principles to practice and clinical applications. Genes Chromosomes Cancer 2018;57:123-39. [Crossref] [PubMed]
- Revelo AE, Martin A, Velasquez R, et al. Liquid biopsy for lung cancers: an update on recent developments. Ann Transl Med 2019;7:349. [Crossref] [PubMed]
- Chan KC, Lai PB, Mok TS, et al. Quantitative analysis of circulating methylated DNA as a biomarker for hepatocellular carcinoma. Clin Chem 2008;54:1528-36. [Crossref] [PubMed]
- Chan KC, Jiang P, Chan CW, et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci U S A 2013;110:18761-8. [Crossref] [PubMed]
- Balgkouranidou I, Chimonidou M, Milaki G, et al. Breast cancer metastasis suppressor-1 promoter methylation in cell-free DNA provides prognostic information in non-small cell lung cancer. Br J Cancer 2014;110:2054-62. [Crossref] [PubMed]
- Tian X, Sun B, Chen C, et al. Circulating tumor DNA 5-hydroxymethylcytosine as a novel diagnostic biomarker for esophageal cancer. Cell Res 2018;28:597-600. [Crossref] [PubMed]
- Chan KC, Jiang P, Zheng YW, et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem 2013;59:211-24. [Crossref] [PubMed]
- Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A 2005;102:16368-73. [Crossref] [PubMed]
- Yung TK, Chan KC, Mok TS, et al. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res 2009;15:2076-84. [Crossref] [PubMed]
- Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013;497:108-12. [Crossref] [PubMed]
- Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012;4:136ra68. [Crossref] [PubMed]
- Leary RJ, Sausen M, Kinde I, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med 2012;4:162ra154. [Crossref] [PubMed]
- Heitzer E, Auer M, Hoffmann EM, et al. Establishment of tumor-specific copy number alterations from plasma DNA of patients with cancer. Int J Cancer 2013;133:346-56. [Crossref] [PubMed]
- Heitzer E, Ulz P, Belic J, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med 2013;5:30. [Crossref] [PubMed]
- Leary RJ, Kinde I, Diehl F, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med 2010;2:20ra14. [Crossref] [PubMed]
- McBride DJ, Orpana AK, Sotiriou C, et al. Use of Cancer-Specific Genomic Rearrangements to Quantify Disease Burden in Plasma from Patients with Solid Tumors. Genes Chromosomes Cancer 2010;49:1062-9. [Crossref] [PubMed]
- Newton CR, Graham A, Heptinstall LE, et al. Analysis Of Any Point Mutation In DNA - the Amplification Refractory Mutation System (Arms). Nucleic Acids Res 1989;17:2503-16. [Crossref] [PubMed]
- Seyama T, Ito T, Hayashi T, et al. A novel blocker-PCR method for detection of rare mutant alleles in the presence of an excess amount of normal DNA. Nucleic Acids Res 1992;20:2493-6. [Crossref] [PubMed]
- Steinman C. Specific inhibition of the polymerase chain reaction using a non-extendable oligonucleotide blocker. US; 1998.
- Nie H, Evans AA, London WT, et al. Ultrasensitive quantification of hepatitis B virus A1762T/G1764A mutant by a SimpleProbe PCR using a wild-type-selective PCR blocker and a primer-blocker-probe partial-overlap approach. J Clin Microbiol 2011;49:2440-8. [Crossref] [PubMed]
- Agarwal P, Kwiatkowski R. Novel acid detection assays employing blocker oligonucleotides 2007.
- Li J, Wang L, Mamon H, et al. Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing. Nat Med 2008;14:579-84. [Crossref] [PubMed]
- Li J, Makrigiorgos GM. COLD-PCR: a new platform for highly improved mutation detection in cancer and genetic testing. Biochem Soc Trans 2009;37:427-32. [Crossref] [PubMed]
- Milbury CA, Li J, Liu P, et al. COLD-PCR: improving the sensitivity of molecular diagnostics assays. Expert Rev Mol Diagn 2011;11:159-69. [Crossref] [PubMed]
- Vargas DY, Kramer FR, Tyagi S, et al. Multiplex Real-Time PCR Assays that Measure the Abundance of Extremely Rare Mutations Associated with Cancer. PLoS One 2016;11:e0156546. [Crossref] [PubMed]
- Milbury CA, Li J, Makrigiorgos GM. Ice-COLD-PCR enables rapid amplification and robust enrichment for low-abundance unknown DNA mutations. Nucleic Acids Res 2011;39:e2. [Crossref] [PubMed]
- Song C, Liu Y, Fontana R, et al. Elimination of unaltered DNA in mixed clinical samples via nuclease-assisted minor-allele enrichment. Nucleic Acids Res 2016;44:e146. [Crossref] [PubMed]
- Hindson CM, Chevillet JR, Briggs HA, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods 2013;10:1003-5. [Crossref] [PubMed]
- Chen WW, Balaj L, Liau LM, et al. BEAMing and Droplet Digital PCR Analysis of Mutant IDH1 mRNA in Glioma Patient Serum and Cerebrospinal Fluid Extracellular Vesicles. Mol Ther Nucleic Acids 2013;2:e109. [Crossref] [PubMed]
- Krug AK, Enderle D, Karlovich C, et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann Oncol 2018;29:2143. [Crossref] [PubMed]
- Metzker ML. Sequencing technologies - the next generation. Nature reviews Genetics 2010;11:31-46. [Crossref] [PubMed]
- Heather JM, Chain B. The sequence of sequencers: The history of sequencing DNA. Genomics 2016;107:1-8. [Crossref] [PubMed]
- Hawkins SF, Guest PC. Multiplex analyses using real-time quantitative PCR. Methods Mol Biol 2017;1546:125-33. [Crossref] [PubMed]
- Han X, Wang J, Sun Y. Circulating tumor DNA as biomarkers for cancer detection. Genomics Proteomics Bioinformatics 2017;15:59-72. [Crossref] [PubMed]
(English Language Editor: K. Brown)


