
Development of a prognostic signature for bladder cancer based on immune-related genes
Introduction
Bladder cancer (BC) is the 9th most common cancer in the United States, with 17,240 estimated new deaths per year (1). In the past 30 years, the prognosis of BC patients has been poor because the diagnosis is often late, and treatment has barely progressed (2). Moreover, patients with the same clinical or pathological stage experience different clinical outcomes, even if patients receive very similar treatments. The genetic heterogeneity of patients contributes the most to their inherent clinical diversity (3). Thus, in the era of precision medicine, molecular biomarkers are urgently needed to estimate prognosis in BC patients and guide precise treatment (4,5).
In the genomics era, molecular biomarkers that can reliably predict patient survival would have important value in the management of several cancers, including BC (6-9). Numerous studies have generated multiple gene expression-based prognostic signatures for patient survival stratification in BC (9,10). However, due to problems such as excessive fitting in the training set and a lack of sufficient validation, none of them have been incorporated into routine clinical practice. Currently, public, and large-scale gene expression data sets are easy to obtain and can provide an opportunity to identify potentially reliable biomarkers for BC patients. However, the diversity of data also represents a daunting challenge. Gene expression levels sequenced by traditional approaches require suitable normalization, which is difficult considering biological heterogeneity and technical biases across microarray and sequencing platforms. Instead, researchers have developed new methods to eliminate the limitations to data processing, such as normalization and scaling based on the relative ranking of gene expression levels, and produced robust results in various studies.
Some components of the immune system have proven to be key factors in the onset and progression of cancer (11-13). The immune checkpoints strictly control immune function to maintain self-tolerance and minimize tissue destruction when an immune response occurs in the surrounding tissues (13-15). Several immune checkpoints have been identified and developed into therapeutic targets for numerous cancers. Recent immunotherapies for immune checkpoints, such as programmed death-1 or programmed death ligand 1, have shown a remarkable, durable response in BC (15-18). Recent emerging evidence has demonstrated that multiple immune gene-based prognostic signatures are potential biomarkers that can estimate overall survival in patients with colorectal, ovarian, and lung cancers (13,19,20). Thus, the immune system-associated gene-based prognostic signature is promising for use in BC. Regarding their prognostic potential in BC, the molecular characteristics of tumor immune interactions remain to be extensively studied.
In our study, to eliminate the requirement for data normalization, using the gene pair method to rank the gene expression levels of microarray and RNA-Seq data sets, we developed and validated a robust and personalized immune prognostic signature. Moreover, to better leverage the complementary role of clinical and molecular markers, a composite prognostic nomogram was constructed by combining the individualized immune-related gene pair (IRGP) signature with the TNM stage, which thus improved the prognostic accuracy for patients with BC.
Methods
Public datasets and study design
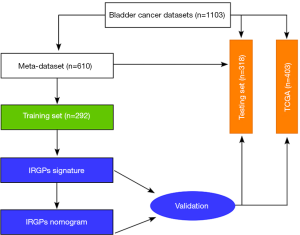
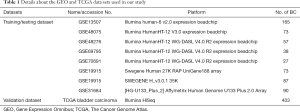
Our study analyzed gene expression in bladder tumor tissue samples obtained from public datasets (Figure 1, Table 1). In total, nine studies were selected, including eight microarray datasets and one RNA-Seq data set. The microarray data and corresponding clinical information were downloaded from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) and normalized with the robust multiarray average (RMA) method, and bias effects across batches were corrected using the Combat method (21). The Cancer Genome Atlas (TCGA) bladder Carcinoma RNA-Seq data set and clinical information were downloaded from the Cancer Genomics Browser (https://genome-cancer.ucsc.edu/) in March 2019. We excluded patients who had received neoadjuvant therapy or other pharmaceutical treatments that may affect the immune system response. In addition, only patients with available and valid survival information were enrolled in all studies. In total, 1,013 patients were included in our study (Table 1). All the microarray datasets were merged as a meta-data set, which was then randomly divided into training and testing datasets according to an approximate 1:1 ratio. No further normalization methods were needed for merging different datasets. The TCGA data set was used as a validation cohort to evaluate the robustness of the IRGP model. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Full table
Identification of BC-specific IRGPs and construction of the prognostic immune signature
We focused on immune-related genes (IRGs) to construct a prognostic immune gene signature. A total of 1,811 unique IRGs were downloaded from the ImmPort database (22) (https://immport.niaid.nih.gov) accessed on March 30th, 2019. These IRGs can be classified into 17 categories, including the B cell antigen receptor signaling pathway, the T cell receptor signaling pathway, natural killer cell cytotoxicity, antigen processing and presentation pathways, and cytokines or cytokine receptors. Only the IRGs measured by all platforms in any data set were selected. Pairwise comparison was conducted for the gene expression level in a specific sample to compute a score for each IRGP. If IRG 1 was less than IRG 2, the output score was 1, while the IRGP score was 0 for the others. After removing IRGPs with constant values (0 or 1, which indicates almost no changes among patients) in any individual data set, the remaining IRGPs were selected as initial candidate IRGPs for the prognostic analysis. The development of a prognostic immune signature was conducted as described previously (13,19). From the initial candidate IRGPs, an IRGP signature was generated using least absolute shrinkage and selection operator (LASSO) Cox proportional hazards regression, and 30 immune gene pairs were used to construct the final prognostic immune model. We determined the optimal cutoff value of the IRGP signature to stratify patients into low- and high-risk groups using a receiver operating characteristic (ROC) curve at 5 years in the training cohort for overall survival. A nearest neighbor estimation method was used to estimate the time-dependent ROC curve. The shortest distance in the ROC curve was used to define the optimal cutoff value. We used CIBERSORT to analyze the infiltration of immune cells in different risk groups.
Validation of the IRGP signature
The prognostic value of the IRGP signature was assessed in the training, testing and independent validation cohorts by the log-rank test. Moreover, we then combined the IRGP signature with the TNM stage in the multivariate analysis. The TNM stage was treated as a continuous variable. Stage I was replaced with 1; stage II was replaced with 2; stage III was replaced with 3; and stage IV was replaced with 4. Patients with T4a and Nx were considered 3.5. When no pathological TNM stage was available, we used the clinical TNM stage instead. The prognostic performance of the IRGP signature was evaluated using the concordance index (C-index), calibration, and decision curve analysis.
Construction and validation of an IRGP nomogram
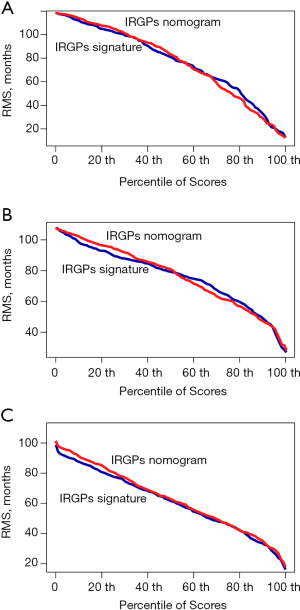
We integrated the TNM stage and the IRGP signature risk score to develop an IRGP nomogram using Cox proportional hazards regression in the training cohort. The prognostic efficiency of the IRGP nomogram was compared with the C-index of the IRGP signature and depicted by the restricted mean survival (RMS) curve, which indicates the life expectancy of BC patients with different risk scores at 10 years.
Functional enrichment analysis
GO annotation (http://www.geneontology.org) and KEGG signaling pathway were set up using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/) bioinformatics tool (23,24).
Statistical analysis
In this study, we performed all statistical analyses using R software (version 3.5.1; https://www.Rproject.org/). Continuous variables were compared using Student’s t-tests or Wilcoxon rank-sum tests. Survival analyses were performed using the Kaplan-Meier method and compared using a log-rank test with the ‘survival’ package. Univariate and multivariate Cox regression analyses were conducted for the IRGP signature and the TNM stage. Time-dependent ROC curve analysis, LASSO Cox regression analysis with 10-fold cross-validation, and nomogram and decision curve analyses were estimated using R packages (survivalROC, glmnet, rms, and DecisionCurve). The C-index was calculated with the R package survcomp and compared with the R package compareC. Unless otherwise stated, statistical significance was set as a P value <0.05.
Results
Discovery of prognostic IRGPs and construction of the IRGP signature
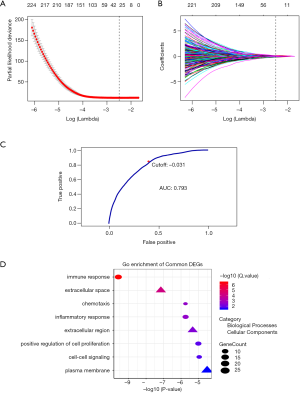
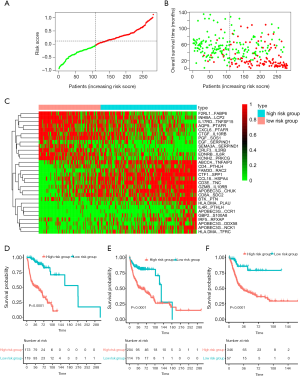
In total, 1,013 patients with BC were enrolled in our study, and the detailed study design is depicted in Figure 1. To enrich the sample number for model development, we combined all the datasets from the GEO, which were obtained from the public website. A total of 610 BC patients with available survival information derived from the GEO were included and then randomly divided into a training cohort (n=292) and a testing cohort (n=318) (Table 1). Among the 1,811 immune-related genes (IRGs) obtained from the ImmPort database, 561 IRGs were detected in all platforms, and 157,080 immune-related gene pairs (IRGPs) were generated. After removing IRGPs with relatively small variation (median absolute deviation =0) in any data set, 44,385 candidate IRGPs were left to further analyze the association between each IRGP and overall survival in BC patients. The clinical characteristics of patients in the training and testing cohorts were well balanced (Table 2). We assessed the association of the candidate IRGPs with overall survival and found 791 prognostic IRGPs with a familywise error rate less than 0.05. Using LASSO Cox regression and 10-fold cross-validation, we then developed a thirty-IRGP signature including 52 IRGs based on the training data set (Figure 2). The 52 different IRGs among the IRGP signature participate in many immune processes, mainly including antimicrobials and cytokines (Table 3). The optimal cutoff value for the IRGP signature was −0.031, which was calculated using time-dependent ROC curve analysis, and stratified BC patients into the low- or high-risk group (Figure 2C). The risk scores of the patients are illustrated in Figure 3, which show the overall survival distribution among different risk groups (Figure 3A,B). The heat map shown in Figure 3C represents the expression levels of the 30 selected gene pairs in the low- and high-risk groups.

Full table
Full table
Functional analysis of the IRGP signature
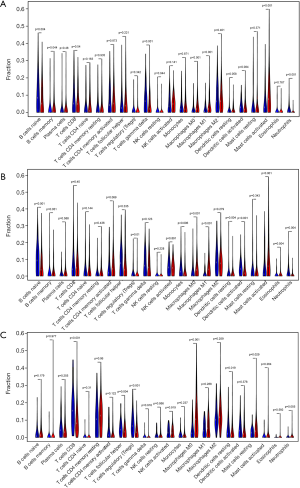
To obtain biological information on the IRGP signature, we carried out an enrichment analysis of the 52 unique IRGs with the DAVID tool. The biological processes of gene ontology with a false discovery rate (FDR) value less than 0.05 were selected, and eight overrepresented biological processes in gene ontology were identified (Figure 2D). Most biological processes were the immune response, chemotaxis, and the inflammatory response. To determine the immune function of the obtained risk groups, we conducted an immune infiltration analysis in the GEO and TCGA datasets using CIBERSORT, which is a popular algorithm describing cell composition from the gene expression profiles of bulk tumors, and found that the percentages of macrophages (M0/M1), activated mast cells and neutrophil infiltration were significantly different between risk groups in the IRGP signature (Figure 4). The mean level of macrophages in the IRGPI high-risk group was 2-fold higher than that in the IRGPI low-risk group in each data set. Patients with a higher IRGPI also had significantly lower B cell naive infiltration in their tumors. However, no statistically significant difference in T cell infiltration was observed between the 2 IRGPI risk groups in the different datasets.
Validation of the prognostic value of the IRGP signature
Based on the defined risk groups, we found that the IRGP signature was significantly associated with overall survival [Figure 3D; HR, 10.91 (6.76–17.60); P=2.0×10−16] in the training cohort. To evaluate whether the IRGP signature had similar prognostic value in different populations, we applied the same formula to the testing and validation cohorts. As expected, the IRGP signature significantly stratified patients in terms of overall survival [Figure 3E; HR, 2.99 (1.92–4.64); P=3.0×10−7] in the testing data set. In the TCGA validation cohort, patients in the high-risk group experienced significantly worse overall survival than patients in the low-risk group [Figure 3F; HR, 4.75 (2.10–10.75); P=4.0×10−5)].
Developing an integrated prognostic nomogram by combining the IRGP signature with clinical features
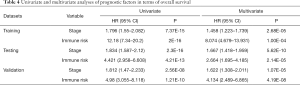
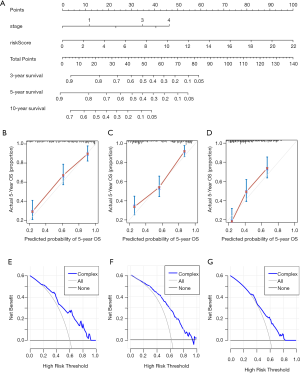
To further evaluate whether the IRGP signature could serve as an independent prognostic factor, univariate and multivariate Cox regression analyses were conducted. In the training cohort, both the univariate and multivariate analyses revealed that the TNM stage and the IRGP signature were significantly associated with overall survival (Table 4). To further improve the predictive accuracy, based on the above results, we developed an IRGP nomogram that combined the TNM stage and the IRGP signature (Figure 5, Table 4). The risk scores of the IRGP nomogram were calculated as (0.378 × stage) + (2.089 × IRGP signature). We calculated the optimal cutoff value of 1.012 based on time-dependent ROC curve analysis in the training set, which was used to divide patients into different prognosis groups. The estimation accuracy of overall survival was enhanced by the continuous form of the IRGP nomogram relative to the IRGP signature (Figure 6, mean C-index, 0.82 vs. 0.79 in the training cohort). In addition, the calibration curves for 5-year overall survival performed well in the training, testing and validation cohorts (Figure 5B,C,D). After a decision curve analysis was conducted for the IRGP nomogram, we also found the model’s clinical application value (Figure 5E,F,G). The results showed that the IRGP nomogram added more net benefit in the testing data set if the threshold probability was between 0% and 92%, while in the training and validation datasets, decision-making based on the IRGP nomogram could add more net benefit than the treating all patients or none scheme if the threshold probability was between 0% and 80%. In summary, our constructed IRGP nomogram is clinically useful for BC patients and may tailor the current treatment strategy.
Full table
Discussion
Patients with BC are at substantial risk for recurrence and metastasis within 5 years after diagnosis (25). The TNM staging system is a common method used to predict the efficacy of systemic therapy and patient survival (26). However, the prognosis of patients at the same stage varies greatly regardless of whether the treatment is similar, which may be due to individual genetic differences (9,10,19). Hitherto, researchers have demonstrated that certain gene expression signatures can estimate survival in BC patients; however, none have been translated into clinical practice (9,10). One common disadvantage of all current experiments is the normalization of gene expression profiles that may produce inherent technical biases across different microarray or RNA-Seq platforms (19).
To screen and develop robust molecular biomarkers in BC, we used a gene pair method that can overcome inherent technological biases between different platforms. Our prognostic immune signature was constructed based on the relative rank of gene expression values involving only a single tumor sample, and data standardization was not needed. Therefore, our prognostic signature may assess patient prognosis in an individualized, single-sample form without reference to other samples and would then be easily adopted clinically. Based on 30 prognostic-associated IRGPs, our developed immune signature reflects distinct biological processes that could function as a marker for estimating patient prognosis in BC. The prognostic immune signature associated with the immuno-microenvironment of tumors may reveal a new prospect for novel predictive biomarker development and improve the efficacy of BC patients in the era of precision medicine. Our developed IRGP signature can stratify patients into low- and high-risk subgroups based on different overall survival rates. In addition, we combined the IRGP signature with the TNM stage, which is currently being used for the staging classification of most cancers, and found that the integrated IRGP nomogram may propose a more accurate estimation of overall survival in BC patients.
Cancer immunotherapy has made great breakthroughs, and IRGs may hold great prospects for identifying new molecular targets (12,27-31). Most genes included in our IRGP signature were antimicrobials and cytokines, which play key roles in the immune response, chemotaxis, and the inflammatory response. Previous studies have shown that adjuvant antimicrobial agents can kill oncogenic-related microorganisms and exert antiproliferative and cytotoxic effects (32-35). Moreover, we found significantly increased infiltration levels of macrophages (M0/M1) and activated mast cells in the immune high-risk group, which were validated in the testing and validation datasets. Based on the results of the current study, immune cell subgroups may play a role in the observed prognostic differences between risk groups defined by our immune signature. The immune genes involved in the IRGP signature have a strong biological background, making it possible for them to be used to guide clinical adjuvant treatment in the future.
Our study still has some limitations. First, the retrospective nature and inherent intratumor genetic heterogeneity in our study may influence the estimation accuracy, although a large number of samples were included for broader validation. Second, our established immune gene signature is based on gene expression produced by microarray and RNA-seq platforms. Due to the high detection costs and expert requirement for bioinformatics analysis, it is difficult to widely generalize the signature in clinical applications. Thus, we need more data from different platforms to validate our developed immune signature.
Conclusions
In summary, our constructed IRGP signature could be used as a potential prognostic biomarker in BC. We also found a positive correlation between the signature and the infiltration of immune cell subsets, such as the immune response, macrophages (M0/M1), and activated mast cells. In the era of immunotherapy, it is necessary to conduct prospective studies to further evaluate the accuracy of predictions and test their clinical application in personalized treatment for BC.
Acknowledgments
We thank the TCGA and GEO Research Network and the involved patients. We apologize for any omission of citations and references due to space limitation.
Funding: Our study was funded by Fundamental Research Funds for the Central Universities of Central South University (grant/award number: 2016zzts121), National Natural Science Foundation of China (grant/award numbers: 81700665, 81572523), and Hunan Province Funds for Distinguished Young Scientists of China (grant/award number: 2016JJ1026).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-1102). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Seiler R, Gibb EA, Wang NQ, et al. Divergent Biological Response to Neoadjuvant Chemotherapy in Muscle-invasive Bladder Cancer. Clin Cancer Res 2019;25:5082-93. [Crossref] [PubMed]
- Sjödahl G, Jackson CL, Bartlett JM, et al. Molecular profiling in muscle-invasive bladder cancer: more than the sum of its parts. J Pathol 2019;247:563-73. [Crossref] [PubMed]
- Margalit O, Mamtani R, Yang YX, et al. Assessing the prognostic value of carcinoembryonic antigen levels in stage I and II colon cancer. Eur J Cancer 2018;94:1-5. [Crossref] [PubMed]
- Hutchinson L. Gastrointestinal cancer: CDX2: prognostic marker for high-risk colon cancer. Nat Rev Clin Oncol 2016;13:134-5. [Crossref] [PubMed]
- Bielli P, Panzeri V, Lattanzio R, et al. The Splicing Factor PTBP1 Promotes Expression of Oncogenic Splice Variants and Predicts Poor Prognosis in Patients with Non-muscle-Invasive Bladder Cancer. Clin Cancer Res 2018;24:5422-32. [Crossref] [PubMed]
- Xie X, Tan W, Chen B, et al. Preoperative prediction nomogram based on primary tumor miRNAs signature and clinical-related features for axillary lymph node metastasis in early-stage invasive breast cancer. Int J Cancer 2018;142:1901-10. [Crossref] [PubMed]
- Gong C, Tan W, Chen K, et al. Prognostic Value of a BCSC-associated MicroRNA Signature in Hormone Receptor-Positive HER2-Negative Breast Cancer. EBioMedicine 2016;11:199-209. [Crossref] [PubMed]
- Zhan Y, Du L, Wang L, et al. Expression signatures of exosomal long non-coding RNAs in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Mol Cancer 2018;17:142. [Crossref] [PubMed]
- Du L, Duan W, Jiang X, et al. Cell-free lncRNA expression signatures in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med 2018;22:2838-45. [Crossref] [PubMed]
- Zhou R, Zhang J, Zeng D, et al. Immune cell infiltration as a biomarker for the diagnosis and prognosis of stage I-III colon cancer. Cancer Immunol Immunother 2019;68:433-42. [Crossref] [PubMed]
- Christopher MJ, Petti AA, Rettig MP, et al. Immune Escape of Relapsed AML Cells after Allogeneic Transplantation. N Engl J Med 2018;379:2330-41. [Crossref] [PubMed]
- Wu J, Zhao Y, Zhang J, et al. Development and validation of an immune-related gene pairs signature in colorectal cancer. Oncoimmunology 2019;8:1596715. [Crossref] [PubMed]
- Friedlander P, Wood K, Wassmann K, et al. A whole-blood RNA transcript-based gene signature is associated with the development of CTLA-4 blockade-related diarrhea in patients with advanced melanoma treated with the checkpoint inhibitor tremelimumab. J Immunother Cancer 2018;6:90. [Crossref] [PubMed]
- Huang J, Shiao SL, Furuya H, et al. Immunogenomic Analysis of Exceptional Responder to ALT-803 (IL-15 Analogue) in BCG Unresponsive Nonmuscle Invasive Bladder Cancer: A Case Series and Review of the Literature. J Immunother 2019;42:354-8. [Crossref] [PubMed]
- Patel KR, Taylor BL, Khani F, et al. Impact of Neoadjuvant Chemotherapy on Concordance of PD-L1 Staining Fidelity between the Primary Tumor and Lymph Node Metastases in Bladder Cancer. Urology 2019;131:150-6. [Crossref] [PubMed]
- Segovia C, San Jose-Eneriz E, Munera-Maravilla E, et al. Inhibition of a G9a/DNMT network triggers immune-mediated bladder cancer regression. Nat Med 2019;25:1073-81. [Crossref] [PubMed]
- Ghate K, Amir E, Kuksis M, et al. PD-L1 expression and clinical outcomes in patients with advanced urothelial carcinoma treated with checkpoint inhibitors: A meta-analysis. Cancer Treat Rev 2019;76:51-6. [Crossref] [PubMed]
- Li B, Cui Y, Diehn M, et al. Development and Validation of an Individualized Immune Prognostic Signature in Early-Stage Nonsquamous Non-Small Cell Lung Cancer. JAMA Oncol 2017;3:1529-37. [Crossref] [PubMed]
- Shen S, Wang G, Zhang R, et al. Development and validation of an immune gene-set based Prognostic signature in ovarian cancer. EBioMedicine 2019;40:318-26. [Crossref] [PubMed]
- Stein CK, Qu P, Epstein J, et al. Removing batch effects from purified plasma cell gene expression microarrays with modified ComBat. BMC Bioinformatics 2015;16:63. [Crossref] [PubMed]
- Bhattacharya S, Andorf S, Gomes L, et al. ImmPort: disseminating data to the public for the future of immunology. Immunol Res 2014;58:234-9. [Crossref] [PubMed]
- Huang W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Huang DW, Sherman BT, Tan Q, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 2007;35:W169-75. [Crossref] [PubMed]
- Soukup V, Capoun O, Cohen D, et al. Prognostic Performance and Reproducibility of the 1973 and 2004/2016 World Health Organization Grading Classification Systems in Non-muscle-invasive Bladder Cancer: A European Association of Urology Non-muscle Invasive Bladder Cancer Guidelines Panel Systematic Review. Eur Urol 2017;72:801-13. [Crossref] [PubMed]
- Lee D, Yoo S, You D, et al. Lymph node density vs. the American Joint Committee on Cancer TNM nodal staging system in node-positive bladder cancer in patients undergoing extended or super-extended pelvic lymphadenectomy. Urol Oncol 2017;35:151.e1-e7. [Crossref] [PubMed]
- Toffalori C, Zito L, Gambacorta V, et al. Immune signature drives leukemia escape and relapse after hematopoietic cell transplantation. Nat Med 2019;25:603-11. [Crossref] [PubMed]
- Yeong J, Lim JCT, Lee B, et al. Prognostic value of CD8 + PD-1+ immune infiltrates and PDCD1 gene expression in triple negative breast cancer. J Immunother Cancer 2019;7:34. [Crossref] [PubMed]
- Cereda M, Gambardella G, Benedetti L, et al. Patients with genetically heterogeneous synchronous colorectal cancer carry rare damaging germline mutations in immune-related genes. Nat Commun 2016;7:12072. [Crossref] [PubMed]
- Prat A, Navarro A, Pare L, et al. Immune-Related Gene Expression Profiling After PD-1 Blockade in Non-Small Cell Lung Carcinoma, Head and Neck Squamous Cell Carcinoma, and Melanoma. Cancer Res 2017;77:3540-50. [Crossref] [PubMed]
- Long J, Wang A, Bai Y, et al. Development and validation of a TP53-associated immune prognostic model for hepatocellular carcinoma. EBioMedicine 2019;42:363-74. [Crossref] [PubMed]
- Razzak M. Bladder cancer: urologists, time to take the helm as antimicrobial stewards. Nat Rev Urol 2013;10:554. [Crossref] [PubMed]
- Shigemura K, Tanaka K, Hamasuna R, et al. Efficacy of Prophylactic Antimicrobial Administration of Tazobactam/Piperacillin for Radical Cystectomy with Urinary Diversion: A Multicenter Study. Urol Int 2019;102:293-8. [Crossref] [PubMed]
- Rodriguez D, Goulart C, Pagliarone AC, et al. In vitro Evidence of Human Immune Responsiveness Shows the Improved Potential of a Recombinant BCG Strain for Bladder Cancer Treatment. Front Immunol 2019;10:1460. [Crossref] [PubMed]
- Kumari N, Agrawal U, Mishra AK, et al. Predictive role of serum and urinary cytokines in invasion and recurrence of bladder cancer. Tumour Biol 2017;39:1010428317697552. [Crossref] [PubMed]


