
Modular assembly–based approach of loosely packing co-cultured hepatic tissue elements with endothelialization for liver tissue engineering
Introduction
In liver tissue engineering, the key to constructing in vitro liver tissue models is to mimic actual cellular microenvironments and produce stable liver-specific functions closely matching those measured in vivo. Hepatocytes are attachment dependent and require sufficient interactions with the surrounding cells and extracellular matrix (ECM) (1). When cultured in a monolayer, hepatocytes lose their normal phenotype and functions (2). Therefore, three-dimensional (3D) cultures of hepatocytes have been developed to form cell aggregates (3). To date, approaches to culturing cell aggregates mainly include the rotary culture system (4), low-attachment culture plates (5), the hanging drop technique (6), micropatterned substrates (7), and microwell arrays (8,9).
However, a simple culture of pure hepatocyte aggregates is not adequate to mimic the liver tissue structure. In fact, the liver is composed of 80% hepatocytes (parenchymal cells) and 20% non-parenchymal cells (10). Typical non-parenchymal cells, such as vascular endothelial cells, fibroblasts, stellate cells, and Kupffer cells, play crucial roles in angiogenesis, tissue regeneration, and cell signaling (11). Therefore, co-culturing hepatocytes with other non-parenchymal cells is essential to mimic the tissue microenvironment as well as promote cell biological functions (12). Conventional Transwell co-culture is often applied in establishing a platform for in vitro hepatotoxicity research and drug screening (13), but it cannot be used to build 3D tissue elements for further assembly. Using a suspension culture, hepatocytes and non-parenchymal cells are expected to self-assemble into heterogeneous aggregates. However, because of the unstable heterotypic interaction in certain types of cells, cells have been observed to lack close contact, resulting in co-culture failures (14). Thus, cell surface modification (15,16) and magnetic microbeads (17) were introduced to control the cell aggregation process manually. These methods usually require specialized materials and devices, and involve exogenous substances, such as synthetic polymers and magnetic microbeads, which may influence normal cell function and have cytotoxic effects.
A biotechnological breakthrough was reported by Kojima et al. (18), who were the first to use methylcellulose (MC) for the rapid production of cellular aggregates. MC absorbs the solvent of the cell suspension and forces the cells to aggregate. Although cell aggregates can be obtained within just several minutes using this technique, it is relatively difficult to separate cell aggregates and high-viscosity MC material. It is of considerable significance that we can rely on cell self-organization to improve the cell-aggregation quality in co-culture. Among the abovementioned non-parenchymal hepatic cells, fibroblasts possess the function of secreting several types of ECM molecules and soluble growth factors (19). It has been demonstrated that fibroblasts stabilize cell−cell interactions and enhance hepatic functions (20-22). Therefore, combining fibroblasts and hepatocytes in co-cultures is usually recommended for obtaining highly functional aggregates without introducing extra materials.
Co-cultured hepatic cell aggregates can be used as tissue elements for large-scale tissue construction in a modular assembly approach. By designing and fabricating individual tissue modules as “building blocks” and assembling them into an integral whole, it is feasible to build scalable and complex tissue constructs with specific biological functions (23,24). Aggregate-based modular assembly uses either homocellular aggregates or co-cultured heterocellular aggregates to build tissue modules. By means of molding and 3D bioprinting approaches, cellular aggregates can be precisely manipulated and self-organized into macrostructures with unique size, shape, and constitution, which are used as manageable units (25). Based on aggregate assemblies, various functional tissue structures have been generated, including cardiac (26), bone (27), hepatic (28), salivary gland (29), thyroid gland (30), and blood vessel (31) tissues.
We cannot ignore that large tissues and organs require a mass of oxygen and nutrient supply, which are delivered by blood vessels in vivo. Similarly, vascularization plays a pivotal role in constructing in vitro large-scale tissues, especially for liver tissue with active metabolism. Vascular endothelial cells, as a type of non-parenchymal cell, are commonly applied in co-culture to form endothelialized aggregates (32). These aggregates are considered to be pre-vascularized microtissues and are packed together for perfusion culture (33). Endothelial cells gradually mature and self-assemble into intricate capillary networks among aggregates. Inamori et al. (34,35) generated hepatocyte aggregates coated with HUVECs and packed them in hollow fibers to assemble functional hepatic tissue. However, due to the dense packing, some adjacent aggregates started to fuse and lost their original boundaries. Partial interstitial spaces were eliminated, thus blocking perfusion flow and leading to massive cell death. In one of their later relevant studies, cell viability in the culture system decreased to less than 60% after just one day (28). Aiming to solve this problem, we proposed the idea of “loosely-packed hepatic tissue elements” in our previous studies (36,37). We used biodegradable scaffold fibers and made a mixture with formed hepatic cell aggregates in a perfusion culture system, which has been proved to be a simple and effective way of maintaining the interstitial space. Thanks to the ameliorative perfusion condition, we implemented a long-term perfusion culture and acquired self-assembled hepatic tissue with high cell retention and enhanced biological functions. However, in one of our former studies (36) we used primary hepatocytes with limited proliferation ability, which hampered the construction of dense liver tissues. As a possible replacement in this study, we used the highly proliferative HepG2 cell line. This line is derived from a hepatocellular carcinoma and has preserved hepatic functions; thus, it seems to be adequate for our purposes. Moreover, our constructed hepatic tissue elements so far have merely consisted of hepatocytes and endothelial cells. To improve cell interaction and hepatic functions, the introduction of fibroblasts in the cell co-culture should be considered.
Therefore, in this study, we introduced the Swiss 3T3 cell line as fibroblasts to generate advanced hepatic tissue elements. By means of a novel polydimethylsiloxane (PDMS)-based oxygen-permeable microwell device (36,38,39), Swiss 3T3 and HUVECs were co-cultured with HepG2 at a high cell density of 4.0×106 cells/cm2 to construct endothelialized hepatic cell aggregates. Aggregates were mixed with biodegradable poly-L-lactic acid (PLLA) fibers in a PDMS-based bioreactor for a 10-day perfusion culture. Aggregates were loosely packed in a bioreactor and achieved 32% in vivo cell density. As advanced hepatic tissue elements, HepG2/Swiss 3T3/HUVECs co-cultured aggregates showed higher aggregation quality, stronger cell proliferation ability, and better liver-specific functions than pure HepG2 aggregates. This work highlights the importance of fibroblasts in co-culturing high-quality hepatic cell aggregates and validates the advantage of the loosely packed mode in aggregate-based modular assembly. Our study also provides guidance for constructing large-scale liver tissue models.
Methods
Cell preparation
The hepatocellular carcinoma HepG2 cell line (RRID: CVCL_0027) was purchased from the Japanese Collection of Research Bioresources (JCRB) and was applied as hepatocytes in this study. HepG2 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, low glucose containing) (10567022, Gibco, Tokyo, Japan) supplemented with 10% fetal bovine serum (FBS) (10099141, Gibco), 1% nonessential amino acid solution (NEAA) (11140, Gibco), 1% antibiotics (15140, Gibco), and 0.5 mM ascorbic acid 2-phosphate (323-44822, Wako, Tokyo, Japan). Swiss 3T3 cells (JCRB, RRID: CVCL_0123) were cultured in high glucose-containing DMEM (10569010, Gibco) and employed as fibroblasts for co-culture with HepG2 cells. HUVECs (C2519A, RRID: CVCL_2959, Lonza, Basel, Switzerland) were maintained in EGM-2 and were harvested for experimental use between passages two and six. All of the cells were incubated at 37 °C in a humidified 5% CO2 atmosphere. Before inoculation into the microwell device, Swiss 3T3 cells were stained with cell membrane labeling reagent PKH 67 (MINI67, Sigma, Tokyo, Japan), and HUVECs were stained with PKH 26 (MINI26, Sigma) for cell distribution observation.
Formation of endothelialized hepatic cell aggregates in the PDMS-based oxygen-permeable microwell device
Figure 1A shows the structure of the PDMS-based oxygen-permeable microwell device used in this study. The fabrication process of the microwell device is detailed in our previous work (36,38,39). Briefly, a 24-well culture plate designed in-house was used as the main body, which consists of a polycarbonate frame, a slice of PDMS sheet, and a stainless bottom frame, stacked and immobilized by screws. PDMS microwell membranes were installed in each of the culture wells in the 24-well plate. The detailed structure of the microwell membranes is shown in Figure 1B. Hexagonal microwells were arranged in a honeycomb pattern on a 1 mm thick circular PDMS membrane. Each chamber of the microwell measured 126 µm in both the inscribed circle diameter and depth for cell inoculation. Supported by the underlying PDMS sheet, microwell membranes enabled direct oxygen diffusion from the bottom of the plate to the cultured cells.
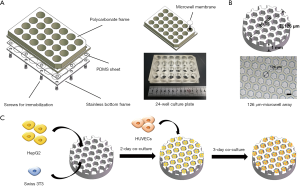
Before cell inoculation, the surfaces of the microwell chambers were treated with a 2-methacryloyloxy ethyl phosphorylcholine (MPC) polymer (lipidure-CR1701, Nichiyu, Japan) coating to prevent cell adhesion (36,38). Cells inoculated in microwells maintained suspension and aggregated spontaneously. As shown in Figure 1C, HepG2 and Swiss 3T3 cells were first seeded at a ratio of 4:1 with a total cell density of 4.0×106 cells/cm2. Pure HepG2 cells at the same cell density were seeded as the control. Twenty hours after inoculation into the microwell device, the aggregate formation of the two groups was evaluated. The average diameter and spheroid circularity of aggregates were calculated by ImageJ (RRID: SCR_003070). After two days of co-culture, HUVECs were subsequently inoculated into microwells at a density of 3.8×104 cells/cm2, and endothelialization on the hepatic cell aggregates was allowed to proceed for another three days of co-culture. Endothelialized hepatic cell aggregates were constructed and harvested for further perfusion culture.
Fiber treatment
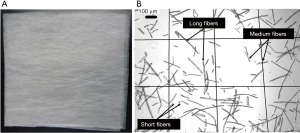
Non-woven fabric (0.1 g, NWF, 20 g/m2) made from PLLA (Figure 2A) was cut into small fragments to serve as biodegradable scaffold fibers. The fibers were treated with 1 N NaOH in an ultrasonic bath for 3 h to enhance their hydrophilicity. After centrifugation, the treated fibers were collected and washed sequentially with 70% ethanol (EtOH), Milli-Q, and phosphate buffer saline (PBS) for three times each. The fibers were then passed through a 300 µm filter to remove fiber clusters. The resulting single fibers (Figure 2B) were dispersed in culture medium for further use.
Perfusion culture
A perfusion bioreactor was fabricated using PDMS for oxygen supply. Figure 3A shows the structure of the bioreactor. The culture medium was dispensed into the culture chamber from the upper flow inlet and discharged from the lower flow outlet. A 40 µm polyester mesh filter was placed at the bottom of the culture chamber to trap cultured elements. The total volume of the culture chamber was 0.1028 cm3. The top tube was designed for aggregate and fiber seeding and was tightly clipped during the perfusion culture.
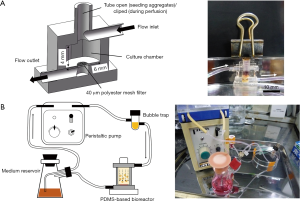
The configuration of the perfusion culture system is illustrated in Figure 3B. The system has four major constituent parts, namely, a medium reservoir, a peristaltic pump, a bubble trap, and a PDMS-based bioreactor. Endothelialized hepatic cell aggregates and PLLA fibers were mixed at a ratio of 1:1 and loaded into the bioreactor. For comparison, pure HepG2 aggregates were used to perform the same perfusion culture. High glucose-containing DMEM was supplied as the perfusion culture medium. The perfusion culture was carried out for 10 days with daily culture medium replacement. During the culture period, the whole perfusion system was placed in an incubator at 37 °C in 5% of CO2. The flow rate of perfusion was controlled at 0.24 mL/min to meet the demand for cellular oxygen consumption.
Immunostaining
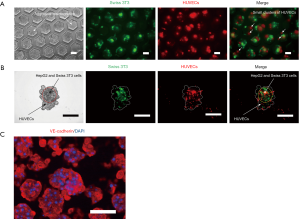
The formed endothelialized hepatic cell aggregates were resuspended and collected from the microwells. The aggregates were fixed with 4% paraformaldehyde for 30 min and permeabilized with 0.1% Triton-X 100 (21123, Sigma) for 20 min, followed by blocking in 1% BSA for another 30 min. Mouse anti-human vascular endothelial (VE)-cadherin (monoclonal, clone BV6, RRID: AB_10845943, MABT134, Merck Millipore, Billerica, MA, USA) was diluted to a ratio of 1:200 and used as a primary antibody. The blocked aggregates were incubated in the primary antibody solution overnight at 4 °C. Subsequently, the aggregates were treated with Alexa 555-conjugated Goat anti-Mouse IgG (polyclonal, IgG(H+L), RRID: AB_2535846, A21425, Invitrogen, USA) as a secondary antibody at a 1:200 dilution for 2 h. Cell nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI) for 15 min. Fluorescent images were captured under a fluorescence microscope (IX 50, Olympus, Tokyo, Japan).
Measurement of DNA content and assessment of liver-specific functions
The bioreactors loaded with aggregates and fibers were fixed in 4% paraformaldehyde solution for 30 min and dehydrated in sucrose solution with gradient concentrations of 10% (2 h), 20% (2 h), and 30% (overnight at 4 °C). The samples were then incubated with PBS containing gelatin for 1 h at 37 °C, and finally blocked at −70 °C. Each sample was vertically cut into two equal parts. One was for DNA content measurement, and the other was for histological morphology observation. For DNA content measurement, the cut samples were sonicated at 40 W for 1 min on ice. The DNA content of suspension was measured by DAPI-fluorometry.
To evaluate the liver-specific functions during the perfusion culture, glucose consumption and albumin production were measured. The culture medium was collected from the medium reservoir and used in a quantitative measurement of glucose concentration using a biochemistry analyzer (YSI 2950, YSI Life Science, OH, USA). To measure albumin production, a sandwich-type enzyme-linked immunosorbent assay (ELISA) was performed using anti-human albumin goat antibody (polyclonal, human albumin, RRID: AB_67016, A80-129A, Bethyl Products, USA) and peroxidase-conjugated anti-human albumin goat antibody (polyclonal, serum albumin, RRID: AB_67023, A80-129P, Bethyl Products, USA) as primary and secondary antibodies, respectively.
Hematoxylin and eosin (H&E) staining
The other part of the divided samples mentioned above was employed in hematoxylin and eosin (H&E) staining. Sections from the central region of each sample were obtained at 10 µm thickness using a microtome and subsequently stained with H&E. The histological morphology of the aggregates was observed under a transmitted light microscope (BX 50, Olympus).
Statistical analysis
All of the data in the results were presented as mean ± standard deviation. A Student’s t-test was used to compare the means of the pure HepG2 group and the co-cultured group. Statistical significance was defined as *, P<0.05, **, P<0.01, and ***, P<0.001.
Results
Co-culture of HepG2 and Swiss 3T3 cells
In our pre-experiment, the inoculation ratio of HepG2 and Swiss 3T3 was investigated. With a higher proportion of Swiss 3T3 cells, aggregation of HepG2 cells was obviously accelerated. However, a high ratio of fibroblasts (50%) could cause excessive cell aggregation, resulting in aggregate movement out of the microwell chambers (data not shown). Therefore, taking into account the aggregate morphology and cell growth condition, the percentage of Swiss 3T3 cells was set at 20%.
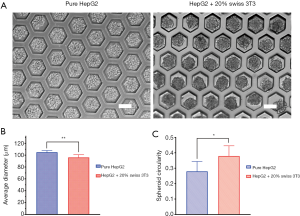
After 20 h of inoculation in the microwell device, compared with pure HepG2 aggregates, HepG2 cells co-cultured with Swiss 3T3 cells formed aggregates with a smoother surface (Figure 4A). The average diameter of co-cultured aggregates was smaller than that of pure HepG2 aggregates (Figure 4B). Co-cultured aggregates showed a higher level of contraction, indicating tighter cell junction. The circularity of the aggregates was also compared in Figure 4C to reflect the aggregate quality. It was noted that co-cultured aggregates had a shape closer to spheroids. This result demonstrated that fibroblasts provide a beneficial function in the formation of hepatic cell aggregates.
Endothelialization of hepatic cell aggregates in the microwell device
On day two after HUVECs inoculation, co-cultured aggregates were observed occupying most of the volume of the microwell chambers. Some overgrown aggregates damaged the MPC coating layer and adhered to the chamber wall, eventually causing aggregate disintegration. Figure 5A illustrates the distribution of Swiss 3T3 cells and HUVECs in the microwells. Interestingly, the seeded HUVECs did not develop a conspicuous cell coating layer as expected. Some HUVECs formed self-aggregated small clusters among the microwell chambers. After filtrating these clusters, detailed fluorescent images of individual aggregates were captured. As shown in Figure 5B, the majority of Swiss 3T3 cells were distributed in the interior region of aggregates, whereas HUVECs were mainly distributed in the peripheral region. As a consequence, a hierarchical structure was constructed with inner HepG2 and Swiss 3T3 cells and outer HUVECs, suggesting the formation of endothelialized hepatic cell aggregates. Figure 5C shows the immunofluorescence of VE-cadherin, which is a specific protein expressed by vascular endothelial cells. As shown in the figure, the co-cultured HUVECs exhibited wide expression of VE-cadherin, indicating the normal biological function of HUVECs in endothelialized hepatic cell aggregates. The results attested to the feasibility of generating functional endothelialized hepatic cell aggregates by means of co-culturing HepG2, Swiss 3T3, and HUVECs in the microwell device.
Fiber preparation
Using the fiber treatment process mentioned above, a fiber suspension with a density of 12 fibers/µL was obtained, which was suitable for mixing with aggregates. The average diameter of fibers was 20 µm, and the length was predominantly in the range of 20−300 µm. Table 1 shows the statistical data of the fiber length distribution in the suspension. Fibers with lengths of 20−50, 50−200, and 200−300 µm were defined as short, medium, and long, respectively. Medium fibers constituted the main part, with a proportion of 43.58%. Short fibers accounted for 28.49%, but were easily washed away through the filter. The remaining 27.93% were made of long fibers, which could break up into shorter segments under the effect of perfused flow.

Full table
Evaluation of cell proliferation and liver-specific functions
The total DNA content of aggregates was measured and normalized to cell number (Figure 6A). On day one of the perfusion cultures, both groups contained approximately 4.0×106 cells. However, after seven days of culture, the two groups exhibited significantly different degrees of cell proliferation. A rapid growth in cell number was observed in co-cultured aggregates, with the value reaching 8.6×106 cells, whereas pure HepG2 aggregates showed a minor proliferation, with 5.8×106 cells. After 10 days of culture, the final cell density of the co-cultured aggregates was 3.24×107/cm3, which is equivalent to 32% of the in vivo cell density (~1×108/cm3). According to this result, cells were able to maintain active proliferation under the co-culture condition.
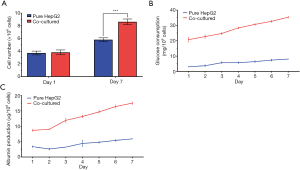
Glucose consumption is relevant for cell respiration and glucose metabolism, which are some of the basic metabolic activities of liver cells. In Figure 6B, the glucose consumption rate of aggregates was tracked during the perfusion culture. For co-cultured aggregates, the amount of glucose consumption maintained a steady augment from 20 mg/106 cells on day one to 35 mg/106 cells on day seven. In contrast, glucose consumption of pure HepG2 aggregates merely showed a slight increase and remained at no more than 10 mg/106 cells. Notably, despite the similar cell numbers between the two groups on day one, the glucose consumption rate of co-cultured aggregates was obviously higher than that of pure HepG2 aggregates. This result was consistent with the result of DNA content measurement, and indicated a stronger proliferation capability and metabolic activity of co-cultured aggregates than pure HepG2 aggregates.
With respect to the assessment of liver-specific secretion function, human albumin production was measured and is shown in Figure 6C. In both groups, albumin production decreased slightly in the first two days and then exhibited steady growth from day three. The reason may be the partial cell loss and the variation of culture conditions. Similarly, albumin secretion of co-cultured aggregates consistently increased, and we noticed a sharper increase during the culture period. On day seven, co-cultured aggregates reached albumin production of 17.5 µg/106 cells, which was a value two times higher than that of pure HepG2 aggregates. On the basis of these results, it can be concluded that co-culturing HepG2 cells with Swiss 3T3 cells and HUVECs enabled the promotion of typical liver-specific functions.
Histological morphology of endothelialized hepatic cell aggregates after perfusion culture
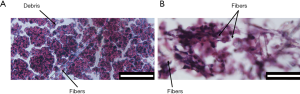
Having accomplished the 10-day perfusion culture, H&E staining was performed to observe the histological morphology of aggregates in the bioreactor. Figure 7 displays images of pure HepG2 aggregates (Figure 7A) and co-cultured aggregates (Figure 7B). Although obvious gaps existed among the pure HepG2 aggregates, some debris separated from the aggregates was observed floating in the interstitial spaces, which could hinder normal perfusion flow. In addition, some of the aggregates converged and lost their original boundaries, leading to the development of non-perfused areas. On the contrary, larger interstitial spaces were retained among co-cultured aggregates. Remarkably, some cells were observed growing out of the co-cultured aggregates. The cells adhered to and spread along the fibers, causing immobilization of the aggregates. Under the effect of cell-fiber interaction, the co-cultured aggregates lost their original spherical shape. Accordingly, aggregates cultured with fibers exhibited a loosely packed structure and generated the necessary spaces for sufficient medium perfusion, which is ideal for long-term culture.
Discussion
Based on a “bottom-up” strategy of tissue engineering, modular assembly provides a promising method of generating large-scale tissues and organoids via fabricating manageable modules and assembling them into a functional whole. Aggregate-based modular assembly uses 3D cultured cell aggregates as functional microtissue modules for later assembly. When endothelialized aggregates are packed together and applied in a perfusion culture system, endothelial cells tend to form intercellular connections that develop into a network structure (34,35,40). However, closely packed aggregates can spontaneously form cell attachments and undergo fusion, leading to the blockage of perfusion flow. Considering this, we added biodegradable PLLA fibers into packed aggregates to acquire the necessary interstitial spaces in normal perfusion culture. Co-cultured aggregates of HepG2/Swiss 3T3/HUVECs were constructed and used as advanced modules for functional liver tissue assembly in this study. The loosely packed aggregates in the bioreactor promoted cell proliferation and liver-specific functions, and finally achieved high cell density, suggesting that this is a feasible approach for constructing large-scale liver tissues.
The co-culture of hepatic cell aggregates was performed in a PDMS-based oxygen permeable microwell device. It is commonly known that hepatocytes consume large amounts of oxygen for metabolic activities. In addition, in view of the density of real liver tissue, aggregates of high cell density should be generated for liver tissue modules. Thus, oxygen supply plays a critical role in aggregate co-culture. Conventional culture plates made of tissue culture-treated polystyrene (TCPS) are not oxygen-permeable. The available oxygen mainly derives from that dissolved in the culture medium. However, due to the poor solubility of oxygen in the culture medium, the amount of cell-acquired oxygen is very limited. To address this problem, we previously fabricated a novel PDMS-based culture plate which contains a PDMS bottom membrane to enable direct oxygen diffusion. The oxygen supply through the PDMS membrane was two orders of magnitude higher than that through the culture medium (41). Based on this difference, we generated high-density cultures of hepatocytes in our previous studies (36,37,39,42). Here, utilizing the same microwell device, we successfully acquired cell aggregates with a cell density of 4.0×106 cells/cm2.
In this study, we applied Swiss 3T3 cells and HUVECs in co-culture with HepG2 cells. Our previous research (36) was based on primary hepatocytes with limited proliferation ability, which is not conducive for the construction of dense liver tissues. Therefore, a type of hepatocellular carcinoma cell (HepG2) with proliferation ability and preserved hepatic functions was employed here. The Swiss 3T3 fibroblast cells synthesize a variety of ECM molecules, such as collagen, elastin, proteoglycan, and fibronectin. HUVECs, as endothelial cells, are essential in angiogenesis for tissue vascularization. Both types of non-parenchymal cells are widely used in the hepatocyte co-culture. In comparison with pure HepG2 aggregates, co-culturing Swiss 3T3 cells with HepG2 cells dramatically facilitated the formation of spherical aggregates. Co-cultured aggregates exhibited a compact cell junction with a smooth surface, which could be related to the ECM secretion by fibroblasts. To endothelialize the aggregates, HUVECs were added to the microwells after two days of HepG2/Swiss 3T3 co-culture. It was noticed that HUVECs formed a non-uniform distribution in the aggregates, and some cells self-aggregated to small clusters. Interestingly, Wenger et al. (43) generated collagen-embedded spheroids by co-culturing HUVECs with human fibroblasts (hFB). Cells self-organized and developed a characteristic spatial organization with HUVECs aggregating in the center and fibroblasts locating peripherally. They deduced that the temporal and spatial organization of co-culture spheroids was highly cell-type dependent, which could be an interpretation to the results of our current study.
Perfusion culture is essential to supply sufficient oxygen and nutrients for cell survival. It has been widely used not only in liver transplantation (44,45) but also in tissue engineering. In our work, during the perfusion culture, HepG2/Swiss 3T3/HUVECs co-cultured aggregates showed a higher level of cell proliferation and liver-specific functions compared with HepG2 aggregates cultured alone. The results concur with those of several relevant studies (20-22). Heterotypic cell−cell interactions existing in co-cultures of hepatocytes with fibroblasts or endothelial cells have been shown to be beneficial to the hepatic phenotype and functional maintenance. Similar results were obtained in co-culturing some other types of non-parenchymal cells, such as stellate cells (46). In our previous work, primary rat hepatocytes were co-cultured with HUVECs (37). Albumin secretion was sharply reduced in the first three days of the perfusion culture, and slightly increased afterward. However, albumin secretion of HepG2/Swiss 3T3/HUVECs co-cultured aggregates only exhibited an unobvious decrease in the early stage, followed by a steady and robust increase. We speculated that hepatic cell aggregates cultured without fibroblasts might lack the necessary ECM components to maintain close cell−cell interactions. The structures of some aggregates eventually collapsed, causing loss of hepatic functions. Thus, our present work highlighted the advantage of co-culturing hepatocytes, fibroblasts, and endothelial cells as a ternary co-culture system.
After 10 days of perfusion culture, we noticed that cells grew out of HepG2/Swiss 3T3/HUVECs co-cultured aggregates and formed adhesion to PLLA fibers, which did not occur in the pure HepG2 aggregates, nor in the previous primary rat hepatocytes/HUVECs aggregates (36). We conclude that Swiss 3T3 cells (fibroblasts) can play an important part in enhancing cell migration. Since HepG2 cells are hepatocellular carcinoma cells, growth factors secreted by fibroblasts such as transforming growth factor-β1 (TGF-β1) (47) may be able to stimulate the metastasis of cancer cells. Fibroblasts could also generate chemokines, such as vascular endothelial growth factor (VEGF), that target endothelial cells (48). Loosely packing aggregates ensured the sufficient supply of culture medium and created an ameliorated mass transfer condition. The long-term culture of aggregates was achieved with good histological morphology. The assembled aggregates reached a cell density of 3.24×107 cells/cm3, which is about 32% that of real tissue. However, further research is still needed. In order to better mimic the biological characteristics of real liver tissue, the cell source should be further considered. Due to the ability of effective expansion in vitro and orientated differentiation, induced pluripotent stem cell (iPS) and embryonic stem cell (ES)-derived hepatocytes exhibit promising potential in liver tissue engineering applications, including the generation of large-scale liver tissue equivalents. To evaluate the effect of tissue vascularization by aggregate assembly, the related biological functions of formed microvascular networks should be further validated. For further in vivo tests, advanced detection methods should be employed to determine the detailed cell composition of formed aggregates, including cell type, number, and ratio. Besides, the mechanism of cell interaction between hepatocytes and other non-parenchymal cells still needs to be clarified, which would help us optimize the cell types and ratios in the aggregate co-culture.
Conclusions
To construct advanced hepatic tissue elements, HepG2 cells, Swiss 3T3 cells, and HUVECs were co-cultured in an oxygen-permeable PDMS-based microwell device. The cell density of aggregates reached 4.0×106 cells/cm2 under sufficient oxygen supply. We found that Swiss 3T3 cells highly promoted the formation of hepatic cell aggregates. The inoculated HUVECs developed a peripheral distribution in the aggregates and supported endothelialization. In comparison to pure HepG2 aggregates, HepG2/Swiss 3T3/HUVECs co-cultured aggregates exhibited a higher level of cell proliferation and expression of liver-specific functions (i.e., glucose consumption and albumin secretion) during the perfusion culture. Biodegradable PLLA scaffold fibers enabled aggregates to pack together loosely, maintaining interstitial spaces for sufficient medium perfusion and thereby ameliorating the mass transfer conditions. A characteristic histological morphology was observed in co-cultured aggregates after 10 days of culture. Some cells migrated out of the aggregates and adhered to the fibers. The final cell density of assembled aggregates in the bioreactor was 3.24×107 cells/cm3, which is nearly 32% of the density of real liver tissue. This study validated the beneficial function of fibroblasts in the hepatic cell co-culture and set the basis for constructing large-scale liver tissue models by the aggregate-based modular assembly.
Acknowledgments
Funding: This work was supported by the National Key Research and Development Program of China (2018YFA0703004), Tsinghua University Initiative Scientific Research Program (20193080045), Tsinghua University-Peking Union Medical College Hospital Initiative Scientific Research Program (20191080843), the National Natural Science Foundation of China (51805294), Tsinghua University Initiative Scientific Research Program (20197050024), and the 111 Project (B17026).
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-1598
Conflicts of Interest: All of the authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-1598). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jain E, Damania A, Kumar A. Biomaterials for liver tissue engineering. Hepatol Int 2014;8:185-97. [Crossref] [PubMed]
- Kyffin JA, Sharma P, Leedale J, et al. Characterisation of a functional rat hepatocyte spheroid model. Toxicol In Vitro 2019;55:160-72. [Crossref] [PubMed]
- van Zijl F, Mikulits W. Hepatospheres: three dimensional cell cultures resemble physiological conditions of the liver. World J Hepatol 2010;2:1-7. [Crossref] [PubMed]
- Tostões RM, Leite SB, Serra M, et al. Human liver cell spheroids in extended perfusion bioreactor culture for repeated-dose drug testing. Hepatology 2012;55:1227-36. [Crossref] [PubMed]
- Raghavan S, Mehta P, Horst EN, et al. Comparative analysis of tumor spheroid generation techniques for differential in vitro drug toxicity. Oncotarget 2016;7:16948-61. [Crossref] [PubMed]
- Hsiao AY, Tung YC, Qu X, et al. 384 hanging drop arrays give excellent Z-factors and allow versatile formation of co-culture spheroids. Biotechnol Bioeng 2012;109:1293-304. [Crossref] [PubMed]
- Uchida S, Itaka K, Nomoto T, et al. An injectable spheroid system with genetic modification for cell transplantation therapy. Biomaterials 2014;35:2499-506. [Crossref] [PubMed]
- Wong SF, No DY, Choi YY, et al. Concave microwell based size-controllable hepatosphere as a three-dimensional liver tissue model. Biomaterials 2011;32:8087-96. [Crossref] [PubMed]
- Zhang B, Li Y, Wang G, et al. Fabrication of agarose concave petridish for 3D-culture microarray method for spheroids formation of hepatic cells. J Mater Sci Mater Med 2018;29:49. [Crossref] [PubMed]
- da Silva Morais A, Vieira S, Zhao X, et al. Advanced Biomaterials and Processing Methods for Liver Regeneration: State-of-the-Art and Future Trends. Adv Healthc Mater 2020.e1901435. [Crossref] [PubMed]
- Godoy P, Hewitt NJ, Albrecht U, et al. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol 2013;87:1315-530. [Crossref] [PubMed]
- Yamada M, Utoh R, Ohashi K, et al. Controlled formation of heterotypic hepatic micro-organoids in anisotropic hydrogel microfibers for long-term preservation of liver-specific functions. Biomaterials 2012;33:8304-15. [Crossref] [PubMed]
- Kostadinova R, Boess F, Applegate D, et al. A long-term three dimensional liver co-culture system for improved prediction of clinically relevant drug-induced hepatotoxicity. Toxicol Appl Pharmacol 2013;268:1-16. [Crossref] [PubMed]
- Powers MJ, Griffith LG. Adhesion-guided in vitro morphogenesis in pure and mixed cell cultures. Microsc Res Tech 1998;43:379-84. [Crossref] [PubMed]
- Kojima N, Takeuchi S, Sakai Y. Establishment of self-organization system in rapidly formed multicellular heterospheroids. Biomaterials 2011;32:6059-67. [Crossref] [PubMed]
- Sasaki K, Akagi T, Asaoka T, et al. Construction of three-dimensional vascularized functional human liver tissue using a layer-by-layer cell coating technique. Biomaterials 2017;133:263-74. [Crossref] [PubMed]
- Desai PK, Tseng H, Souza GR. Assembly of Hepatocyte Spheroids Using Magnetic 3D Cell Culture for CYP450 Inhibition/Induction. Int J Mol Sci 2017;18:1085. [Crossref] [PubMed]
- Kojima N, Takeuchi S, Sakai Y. Rapid aggregation of heterogeneous cells and multiple-sized microspheres in methylcellulose medium. Biomaterials 2012;33:4508-14. [Crossref] [PubMed]
- Schultz GS, Wysocki A. Interactions between extracellular matrix and growth factors in wound healing. Wound Repair Regen 2009;17:153-62. [Crossref] [PubMed]
- Otsuka H, Sasaki K, Okimura S, et al. Micropatterned co-culture of hepatocyte spheroids layered on non-parenchymal cells to understand heterotypic cellular interactions. Sci Technol Adv Mater 2013;14:065003. [Crossref] [PubMed]
- Lu HF, Chua KN, Zhang PC, et al. Three-dimensional co-culture of rat hepatocyte spheroids and NTH/3T3 fibroblasts enhances hepatocyte functional maintenance. Acta Biomater 2005;1:399-410. [Crossref] [PubMed]
- Chia SM, Lin PC, Yu HT. TGF-beta 1 regulation in hepatocyte-NIH3T3 co-culture is important for the enhanced hepatocyte function in 3D microenvironment. Biotechnol Bioeng 2005;89:565-73. [Crossref] [PubMed]
- Schon BS, Hooper GJ, Woodfield TBF. Modular tissue assembly strategies for biofabrication of engineered cartilage. Ann Biomed Eng 2017;45:100-114. [Crossref] [PubMed]
- Ouyang L, Armstrong JPK, Salmeron-Sanchez M, et al. Assembling Living Building Blocks to Engineer Complex Tissues. Adv Funct Mater 2020. [Crossref]
- Lei J, Murphy WL, Temenoff JS. Combination of heparin binding peptide and heparin cell surface coatings for mesenchymal stem cell spheroid assembly. Bioconjug Chem 2018;29:878-84. [Crossref] [PubMed]
- Jakab K, Norotte C, Damon B, et al. Tissue engineering by self-assembly of cells printed into topologically defined structures. Tissue Eng Part A 2008;14:413-21. [Crossref] [PubMed]
- Ahmad T, Byun H, Lee J, et al. Stem cell spheroids incorporating fibers coated with adenosine and polydopamine as a modular building blocks for bone tissue engineering. Biomaterials 2020;230:119652. [Crossref] [PubMed]
- Okudaira T, Amimoto N, Mizumoto H, et al. Formation of three-dimensional hepatic tissue by the bottom-up method using spheroids. J Biosci Bioeng 2016;122:213-8. [Crossref] [PubMed]
- Ozdemir T, Srinivasan PP, Zakheim DR, et al. Bottom-up assembly of salivary gland microtissues for assessing myoepithelial cell function. Biomaterials 2017;142:124-35. [Crossref] [PubMed]
- Bulanova EA, Koudan EV, Degosserie J, et al. Bioprinting of a functional vascularized mouse thyroid gland construct. Biofabrication 2017;9:034105. [Crossref] [PubMed]
- Norotte C, Marga FS, Niklason LE, et al. Scaffold-free vascular tissue engineering using bioprinting. Biomaterials 2009;30:5910-7. [Crossref] [PubMed]
- Sumi S. Self-condensation culture for vascularized organoid. Ann Transl Med 2018;6:S15. [Crossref] [PubMed]
- Mironov V, Visconti RP, Kasyanov V, et al. Organ printing: Tissue spheroids as building blocks. Biomaterials 2009;30:2164-74. [Crossref] [PubMed]
- Inamori M, Mizumoto H, Kajiwara T. An approach for formation of vascularized liver tissue by endothelial cell–covered hepatocyte spheroid integration. Tissue Eng Part A 2009;15:2029-37. [Crossref] [PubMed]
- Inamori M, Mizumoto H, Kajiwara T. Investigation of medium perfusion through scaffold-free tissue constructs using endothelial cell-covered spheroids in vitro. Biochem Eng J 2010;50:116-21. [Crossref]
- Pang Y, Montagne K, Shinohara M, et al. Liver tissue engineering based on aggregate assembly: efficient formation of endothelialized rat hepatocyte aggregates and their immobilization with biodegradable fibres. Biofabrication 2012;4:045004. [Crossref] [PubMed]
- Pang Y, Horimoto Y, Sutoko S, et al. Novel integrative methodology for engineering large liver tissue equivalents based on three-dimensional scaffold fabrication and cellular aggregate assembly. Biofabrication 2016;8:035016. [Crossref] [PubMed]
- Shinohara M, Kimura H, Montagne K, et al. Combination of microwell structures and direct oxygenation enables efficient and size-regulated aggregate formation of an insulin-secreting pancreatic β-cell line. Biotechnol Prog 2014;30:178-87. [Crossref] [PubMed]
- Shinohara M, Komori K, Fujii T, et al. Enhanced self-organization of size-controlled hepatocyte aggregates on oxygen permeable honeycomb microwell sheets. Biomed Phys Eng Express 2017;3:045016. [Crossref]
- Yajima Y, Lee CN, Yamada M, et al. Development of a perfusable 3D liver cell cultivation system via bundling-up assembly of cell-laden microfibers. J Biosci Bioeng 2018;126:111-8. [Crossref] [PubMed]
- Komori K, Udagawa M, Shinohara M, et al. Formation and harvesting of thick pancreatic β-cell sheets on a highly O2-permeable plate modified with poly(n-isopropylacrylamide). Biomater Sci 2013;1:510. [Crossref] [PubMed]
- Evenou F, Hamon M, Fujii T, et al. Gas-permeable membranes and co-culture with fibroblasts enable high-density hepatocyte culture as multilayered liver tissues. Biotechnol Prog 2011;27:1146-53. [Crossref] [PubMed]
- Wenger A, Kowalewski N, Stahl A, et al. Development and characterization of a spheroidal co-culture model of endothelial cells and fibroblasts for improving angiogenesis in tissue engineering. Cells Tissues Organs 2005;181:80-8. [Crossref] [PubMed]
- Schlegel A, Muller X, Dutkowski P. Machine perfusion strategies in liver transplantation. Hepatobiliary Surg Nutr 2019;8:490-501. [Crossref] [PubMed]
- Zhao Q, Nie Y, Guo Z, et al. The future of organ-oriented research and treatment. Hepatobiliary Surg Nutr 2019;8:502-5. [Crossref] [PubMed]
- Lee SA, No DY, Kang E, et al. Spheroid-based three-dimensional liver-on-a-chip to investigate hepatocyte–hepatic stellate cell interactions and flow effects. Lab Chip 2013;13:3529. [Crossref] [PubMed]
- Gupta A, Budhu S, Merghoub T. One checkpoint may hide another: inhibiting the TGFβ signaling pathway enhances immune checkpoint blockade. Hepatobiliary Surg Nutr 2019;8:289-94. [Crossref] [PubMed]
- Sullivan KM, Kenerson HL, Pillarisetty VG, et al. Precision oncology in liver cancer. Ann Transl Med 2018;6:285. [Crossref] [PubMed]


