
Microbiome of the lower genital tract in Chinese women with endometriosis by 16s-rRNA sequencing technique: a pilot study
Introduction
Endometriosis is known as a benign estrogen-depend gynecological disease that mainly affects women in reproductive age, and prevails in 2–10% of the population (1). It is characterized by the growth of endometrial glands and stromal cells inside and outside the pelvic cavity (2-4). The symptom of endometriosis mainly presents with infertility, dysmenorrhea, chronic pelvic pain and dyspareunia (5), and it also places a large burden on the health care system (4). There are three main types of endometriosis: ovarian endometriosis, deep infiltrating endometriosis (DIE), and peritoneal endometriosis. The other special types of endometriosis merely account for a small portion, but still significantly contributes to low quality of life. DIE contributes the most in pelvic pain syndrome, while ovarian endometriosis mainly presents with a pelvic mass (6). Although retrograde menstruation is the most acceptable theory at present, the etiology of endometriosis remains unclear after many researches for over a century. Oxidative stress and genetic features are the other possible important factors that lead to endometriosis (7), and these may affect the immune system and contribute to the growth of endometrial tissue outside the uterine cavity. Many different immunological cells are associated with endometriosis (2), which include macrophages (8), mast cells (9), neutrophils, dendritic cells and gliocytes (2). Some immunological factors, such as interleukins (IL), interferons (IFN), tumor necrosis factors (TNF) and macrophage inflammatory protein (MCP) (7,10), are also correlated to endometriosis (2). Nevertheless, endometriosis is not only a regional restricted disease, but also a systemic immunological dysfunction disease. In clinic, it remains as a challenge to clearly diagnose endometriosis through only non-invasive methods. Ultrasound, magnetic resonance image (MRI) and carbohydrate antigen 125 (CA125) blood test are the recommended methods to evaluate the disease (7), while laparoscopic surgery and pathological examination are the gold standards of diagnosis. To date, gynecologists are short of methods to diagnose endometriosis in early stage. Hence, it is imperative to establish a non-invasive diagnostic model to evaluate and carry out early interventions.
According to a previous study, immunological and genetic function disturbance may increase the munity of bacterial vaginosis (BV) through interleukin and CD4 cell overexpression (11). BV is also associated with higher numbers of chemokine receptor 5 (CCR5) cells, the CD4/CD8 rate, and the interleukin 1β (IL-1β) and tumor necrotic factor-α (TNF-α) level in vaginal-cervical mucosa (12). In healthy women, the vaginal microbiota is dominated by Lactobacillus, and the low-pH environment is maintained by secreting lactate (13,14). The women homozygous for the wild type allele of IL-1ra gene showed increased level of IL-1β when gram negative rods present compared with heterozygous women. Women with an IL-6 gene polymorphism associated with less cytokine response had a higher prevalence of BV, while women with IL1b and IL8 gene polymorphisms associated with increased response had lower prevalence of BV (13). The dysfunction of immunological factors not only increase the risks of human papillary virus/human immunodeficiency virus (HPV/HIV) infection, but also increase the risk of bacterial infection (13,15). Sufficient evidences are available on the micro-environment in the cervical-vaginal region, but it remains debatable whether the uterine cavity is sterilized. Khan et al. validated that lipopolysaccharide was detectable in the endometrium, and that it was highly correlated to endometriosis (16). Other researchers have also successfully verified that the uterine cavity was non-sterilized (17,18) through 16S ribosomal-RNA gene sequencing and bacterial culture (19). However, it remains uncertain whether these positive results were caused by accident contamination from the cervical-vaginal canal during sample collection, or by some unknown etiologies, such as pregnancy (20) and sub-clinical infection (18,19).
A ‘second human genome project’ was proposed in 2001 to explore the human microbiome composition, and investigate the interactions between the microbiome and human host (21). Two methods were mainly used to analyze the microbiome composition, which include culture-based technology and sequencing-based technology. The low throughput culture-based method could not detect the whole microbes of one specific site. The 16S ribosomal-RNA gene sequencing, which is a kind of next-generation high throughput DNA sequencing technology, could make up the deficiency of the culture-based method, and has become the specific standard approach to identity the microbiome (22).
To our knowledge, few relevant studies that simply mentioned the different microbiome composition of the reproductive duct between patients with or without endometriosis by performing 16s-rRNA gene amplicon sequencing (17,18) were found in the systematic review. The present study is aimed to establish a microbiota profile model of endometriosis through the 16s-rRNA gene amplicon sequencing technique and explore significant microbiota in endometriosis or adenomyosis condition.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-1309).
Methods
Patients and sample collection
The Institutional Review Board at the Peking Union Medical College Hospital approved the study (approval No. JS-1532). Informed consent was obtained from all patients, and the study was conducted in accordance with the Declaration of Helsinki Principles (as revised in 2013) and regulations of our institute.
The outpatients and inpatients, who were ready to undergo pre-operational preparation in Peking Union Medical College Hospital from April 2018 to February 2019, were included in the present study. The inclusion criteria included the following items: patients within 18–45 years old, patients with regular menstrual cycle (28±7 days), non-antibiotics use within 30 days, no hormone replacement therapy for 30 days, no sexual activity within 48 hours, no douching and vaginal medications within five days, and no cervical treatment within one week. The exclusion criteria included the following items: patients with BV, cervical inflammation, pelvic inflammatory disease, any acute inflammation, cancer and autoimmune disorders, pregnant patients, patients with intra-uterine devices, and patients who were in menstrual period at the time of sampling. Samples were collected from the cervix on the day of clinical visit for outpatients or before pre-operational vaginal douching for inpatients. Disposable swabs (Jiangsu Tianli Medical Instrument Co., Ltd.) were used for sampling the cervical canal (A) and posterior fornix (B), and vacuum suck tubes (Jiangsu Tianli Medical Instrument Co., Ltd., Guardking, JDC-II) were used for sampling the endometrium (C). In order to minimize the contamination from the cervical canal and vagina, the cervical canal was sterilized with iodine, and the vacuum suck tube was inserted into the uterine cavity to avoid any contact with the vaginal wall. Then, the samples were placed in ice, stored at −80 °C, and transported on dry ice to Annoroad Gene Technology Co. Ltd. (Beijing, China) for further analysis.
The 16S ribosomal-RNA gene sequencing
Total genome DNA was extracted from the samples using the CTAB/SDS method. The DNA concentration and purity were monitored on 1% agarose gels. According to the concentration, the DNA was diluted to 1 ng/µL using sterile water. Then, the 16s-rRNA genes of distinct regions (16SV3-V4) were amplified using specific primers 338F (5'-ACTCCTACGGGAGGCAGCAG-3') and 806R (5'-GGACTACHVGGGTWTCTAAT-3') with the barcode. All PCR reactions were carried out with the Phusion® High-Fidelity PCR Master Mix (New England Biolabs). A certain volume of the 1X loading buffer (containing SYBR green) was mixed with the PCR products, and electrophoresis was performed on 2% agarose gel for detection. Samples with a bright main strip between 400–450 bp were chosen for further experiments. The PCR products were mixed in equidensity ratios. Then, the mixture of PCR products was purified using the Qiagen Gel Extraction Kit (Qiagen, Germany). The sequencing libraries were generated using a TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, USA), according to manufacturer’s recommendations, and index codes were added. The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. Lastly, the library was sequenced on the lluminaHiSeq2500 platform, and 250 bp paired-end reads were generated.
Bioinformatic analysis
Microbiota diversity analysis
The raw sequence reads of the 16s-rRNA gene sequences were spliced, cut, quality filtered and analyzed using the vsearch and usearch tool (23,24) by Terminal in Rstudio. In order to enhance the sensitivity of species recognition, traditional operational taxonomy units (OTU, 97% cluster) was performed to analyze the different bacterial profile between cervical canal and posterior fornix. In order to identify the presentative functions in different disease groups and biomarkers, amplicon sequence variants (ASV, 100% cluster) were taxonomically classified through Unoise3 (25), which is higher in specificity, and using the Greengenes 16s-rRNA gene reference database. The taxonomic composition of microbial communities was visualized using R and STAMP v.2.1.3. Alpha rarefaction curve was conducted by usearch tool in Linux environment. The alpha diversity was computed in four different methods, including the Chao1 index, Richness index, Shanoon_e index and Simpson index. This alpha-rarefaction curve is aimed to estimate whether sample size is big enough to conduct alpha diversity analysis. The community clustering was measured by Bray-Curtis matrix, Unifrac matrix and their corresponding weighted algorithm, based on the normalized OTU table. Principal coordinate analysis (PCoA) and Principal component analysis (PCA) were analyzed by Adonis test, P<0.05 is considered as statistically different.
Biomarker identification
In order to identify the distinct cervical canal microbiota between different groups and explore the biomarker bacterial, the Linear Discriminative Analysis Effect Size (LEfSe) method was used to compare the composition of the cervical canal microbiota using an online tool (http://www.ehbio.com/ImageGP/index.php/Home/Index/LEFSe.html).
Analysis of functional profiles
After filtering and the analysis of OTUs, PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) (25) was performed to determine the different functions of the vaginal bacteria. The vsearch (23) tool was used to analyze the four different groups of participants, and the KEGG Orthology (KO3) functional profiling of the vaginal microbiota was performed. The matrix was normalized by dividing the absolute amount of each functional gene by the total number of reads assigned to the functional genes in each sample (each sample was normalized for 3,900). Merely statistically significant results (P<0.05) were presented in the figures using STAMP v2.1.3.
Results
Samples and participant characteristics
A total of 68 patients were included in the present study, cervical canal (A) and posterior fornix (B) samples were collected from each participant. Among these samples, 67 samples obtained from the cervical canal (A) and 65 samples obtained from the posterior fornix (B) successfully proceeded to the PCR. Due to the informed consent obtained from each patient, merely two samples were collected from the uterine cavity (C) and successfully proceeded to the PCR. Hence, a total of 134 samples were available. Since 15 out-patients did not receive surgical interventions, their final diagnoses were obtained from the radiological examinations, which included their trans-vaginal ultrasound and pelvic magnetic resonance imaging (MRI). A total of 53 participants underwent surgical intervention, and their medical records, including surgery records and pathological reports, were available. Furthermore, 19 (27.94%) participants were diagnosed with adenomyosis, 20 (29.41%) participants were diagnosed with endometriosis (including ovarian endometriosis, DIE, peritoneal type and other special types), 7 (10.29%) participants were diagnosed with adenomyosis accompanied with endometriosis, and 36 (52.94%) patients were classified as control group, which included infertility (without endometriosis verified by laparoscope), leiomyoma, ovarian borderline tumor and teratoma. Among the 134 samples, 25 samples were assigned to the endometriosis (EM) group, 67 samples were assigned to the control (CT) group, 14 samples were assigned to the adenomyosis-endometriosis (AMEM) group, and 28 samples were assigned to the adenomyosis (AM) group. The distribution of sample location and diseases are shown in Table 1. Demographic data is shown in Table 2.

Full table

Full table
The 16s-rRNA gene sequencing of the eligible 67 cervical canal samples, 65 posterior fornix samples and two endometrial samples yielded a total of 7.35 million raw sequences, with 30,084 to 173,739 sequences per sample, and the average length of sequence for each sample ranged within 417.21 to 499.99 bp (Table S1). According to the alpha rarefaction curve estimation, the number of sequences can well represent the microbial diversity of each community (Figure S1).
Microbiota differences in different sites in genital tract
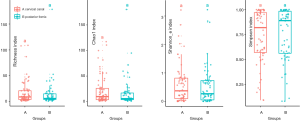
In order to evaluate the different microbiota in different locations in the cervix, PCA and PCoA was conducted. The investigators attempted to determine whether the other sites near the cervix could take the place of the microbiota in the uterine cavity because of the high contamination risk. Two locations near the cervix were analyzed, as previously mentioned (cervical canal and vaginal posterior fornix). ASV was used and all three levels (order, family and genus) of microbiota revealed no significance in the PCA plot (Figure S2). There was no significance found in alpha diversity (Figure S3) and PCoA (Figure S4) (P>0.05). Next, in order to promote the sensitivity, traditional OTU was used to identify the different profile of microbiota between two sample locations. Also, the result revealed no significance in alpha diversity (Figure 1) and PCoA (Figure 2). The proportion of taxa in different locations were shown in boxplot (Figure S5). LeFSe did not identify any significant biomarker between cervical canal and posterior fornix.

Microbiota composition of different diseases
In order to determine whether the composition of microbiota differ, an alpha diversity analysis was conducted. Four groups of boxplots were generated, and no statistical significance was found in alpha diversity (Figure S6).
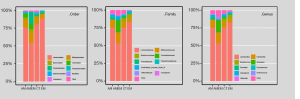
In addition, a beta diversity analysis was also performed to determine the proportion of each OTU, and determine whether the differences were significant. As shown in Figure 3, the analysis was conducted in three different levels. Lactobacillus was the dominant genus in the vagina, as previously reported (11,18). For patients with adenomyosis-endometriosis, in terms of order level, Coriobacteriales shared the largest proportion in the four groups. Coriobacteriaceae was more dominant than any of the other three groups in the family level. In the genus level, Atopobium was greater than any of the other groups.
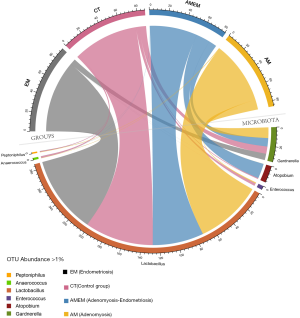
After filtering the raw data, the microbiota with an OTU abundance of >1% was selected in all four groups of participants. Microbiotas were filtered and 6 kinds of bacterial genus were presented in the circular plot (Figure 4). Lactobacillus was the most prevalent microbiota, and the four groups of participants shared this, which was almost the same. However, a large proportion of Atopobium was taken by the AMEM group, while a small proportion was occupied by the other three groups.
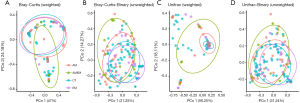
Four PCoA matrix distances were analyzed (Bray-Curtis, Bray-Curtis-Binary, Unifrac, Unifrac-Binary), as shown in Figure 5. The distribution of these four disease groups were not fully separated, but AMEM group is significantly different from others. The p values between each group are shown in Table 3.

Full table
Visualized bar plots were used to explore the different abundance of microbiota with statistical significance (P<0.05). The OTU data were analyzed in the genus and family levels, hoping to determine the quantitative differences among the four groups of participants (Figure 6). In the genus level, four genes of microbiota revealed a higher abundance in the AMEM group, when compared to any of the other three groups. Furthermore, Atopobium, Campylobacter, Ezakiella, Faecalibaterium and Escherichia/Shigella exhibited a higher abundance in the AMEM group, when compared to the CT group and EM group. Atopobium, Campylobacter, Ezakiella, Faecalibaterium are also higher in the AMEM group, when compared to the AM group but Escherichia/Shingella did not show significance between AMEM and AM. In the family level, merely Coriobacteriaceae and Campylobacteriaceae was significantly higher in AMEM group, when compared to any of the other three groups.

The LefSe analysis failed to identify biomarker microbiota (ASV). After the pairwise comparison, no discriminative species were found among the four groups.
Phylogenetic investigation of communities by reconstruction of unobserved states
After the analysis of the different compositions of microbiota among the four groups of diseases, another important question is why does these significant differences exists, and does these contribute to some unknown etiologies of diseases? PICRUSt was performed to explore the different functions of different microbiota (26,27). Furthermore, KO3 functional profiling was performed in two-two comparison (Figure S7).
In the AMEM and EM groups (Figure S7A), 65 KO3 functions were labeled with statistically significance (P<0.05), and nearly half of these were in a small portion of less than 1%. Hence, it is hard to explain the real impact and significance to the diseases.
In the AMEM and CT groups (Figure S7B), 60 KO3 functions were identified (P<0.05).
In the AMEM and AM groups (Figure S7C), 24 functions with statistical significance were identified.
In the AM and CT groups (Figure S7D), 10 KO3 functions were labeled.
In the CT and EM groups (Figure S7E), 20 KO3 functions were identified.
Finally, in the AM and EM groups (Figure S7F), merely metabolism/lipid metabolism/synthesis and degradation of ketone bodies was identified, and this was significantly higher in the EM group than in the AM group.
Overall, several functions overlapped in the two-two comparison. The AMEM group had a greater proportion of “environmental information processing/signal transduction/two-component system, genetic information processing/transcription/transcription factor, genetic information processing/translation/ribosome, and metabolism/glycan biosynthesis and metabolism/peptidoglycan biosynthesis”. However, the EM group had a higher proportion of metabolism/lipid metabolism/synthesis and degradation of ketone bodies.
Discussion
Endometriosis is a disease that involves endometrium tissues and immune system dysfunction (2). After the collection of samples from the uterine cavities, the outcome of the PCR was barely satisfactory, when compared to samples obtained from the cervix. No valid evidence demonstrated that the uterine cavity is sterilized, since the immune system can still affect the micro-environment in the uterine cavity (8,16). Some researchers have successfully proven the non-sterilized environment through 16s-rRNA sequencing (17) and bacterial culture (16) methods. However, it remains uncertain whether other unknown sub-clinical infections or other underlying diseases can affect the environment in the uterine cavity. The “bacterial contamination hypothesis” was proposed by Khan et al. (16). This hypothesis suggests that the lipopolysaccharide level of Escherichia coli is significantly higher in endometriosis patients, and that this significantly contributes to the progression of endometriosis. In addition, Takebayashi et al. suggested that endometriosis is strongly associated with endometritis (28,29). From another aspect, the mucosa of the vagina and cervix are enriched with bacteria, and it is possible for lipopolysaccharides or bacterial reflux to and be present in the uterine cavity and cause immunological reactions. In the present study, two different sites near the cervix (cervical canal A and posterior fornix B) were compared, and an attempted was made to identify a marker site that was able to replace the endometrium environment. However, the available samples obtained from the uterine cavity was not enough, and it was difficult to compare the OTUs with the cervix samples. Furthermore, after the comparison of microbiota between A and B, no statistical differences were obtained through the PCA plots, similar result was reported by Chen et al. (18). This result suggests that the microbiota obtained from different locations near the cervix was not a distinctive marker site. Considering the algorithm and cluster methods during the analysis, ASV is 100% cluster and it has a higher specificity but the sensitivity is comparatively lower than traditional OTU (97% cluster). In this study, since the aim was to find out the different profile of microbiota between posterior fornix and cervical canal, these two locations are also anatomically closed, 97% cluster OTU was attempted after the negative result from ASV, but 97% cluster were still failed to reveal the difference. In this study, the small sample size and the individual difference may result in these insignificances.
The association of adenomyosis and endometriosis had not been clearly demonstrated (30). At present, there are short of studies on 16s-rRNA sequencing in adenomyosis. Hence, adenomyosis patients were included, and an attempted was made to determine the underlying associations through 16s-rRNA sequencing. Four types of index in alpha diversity was conducted: Chao1 index, Richness index, Shanoon_e index and Simpson index. However, no statistical significance was found. This result suggests that endometriosis and adenomyosis may not differ in the richness of microbiota. In addition, all four groups of participants exhibited no significances in alpha diversity. Furthermore, the numbers of existing OTUs were stable, and the composition of the microbiota has an unclear impact to the disease, according to the present results. Hence, more cases should be included and more studies should be conducted to establish a better theory of microbiota relationship. PCoA in beta diversity showed us that adenomyosis-endometriosis is different from control group and the beta diversity revealed a possible result that adenomyosis-endometriosis patients may be correlated to Atopobium, and that Atopobium was more abundant in adenomyosis-endometriosis patients than in any other groups. Atopobium has been previously reported as a BV-related microbiota (31), and that it also colonizes in healthy women (32). Some sub-clinical infections may still exist, although BV was excluded in these present participants through verbal confirmation and microbial nuclei acid detection (Candida Albican, Trichomonas vaginalis and Gardnerella). Previous studies have also demonstrated that endometritis is strongly correlated to endometriosis (28,29). The “bacterial contamination hypothesis” demonstrated that lipopolysaccharide is detected in the endometrium and activates the lipopolysaccharide/Toll like receptor 4 (LPS/TLR4) cascade in women with endometriosis (16). Streptococcaceae, Staphylococaceae and Enterobacteriaceae were significantly elevated in gonadotropin-releasing hormone agonist (GnRHa)-treated women with endometriosis (17,19). These evidences may provide some information that endometriosis is possibly correlated to bacterial contamination, and the present study supports this hypothesis, to a certain extent. However, it remains to be determined which kind of bacteria contributes the most to endometriosis, and how these infect the genital tract. According to the present results, Atopobium was statistically significant in adenomyosis-endometriosis patients, which was opposite to the result reported by Walther-António et al., this difference may result from the different stage of endometriosis and underlying adenomyosis condition (33). Atopobium is carcinogenic, and it can facilitate infection through Porphyromonas species, which can intracellularly manifest and disrupt cell regulatory functions, leading to a carcinogenic trigger (33). Furthermore, it also relates to higher IL-1β levels (13). A previous study also revealed that high IL-1β levels are associated with higher risk of endometriosis (34). It was assumed that adenomyosis and endometriosis may correlate to Atopobium through IL-1β. However, it remains to be determined how the relationship of adenomyosis and endometriosis in the microbiome pathway can be explain through 16s-rRNA sequencing. the present study suggests that Atopobium may be involved in the progression of endometriosis through certain unknown mechanisms, such as IL-1β and Atopobium infection.
PICRUSt revealed a novel concept that endometriosis and adenomyosis may be associated with certain mechanisms through a predictive microbiota function analysis. Ribosome is a function that should be noticed, and previous study on ribosomes suggested that the upregulation of ribosomes may promote the progression of endometriosis (35). However, there are no reports on the association of adenomyosis and ribosomes, to date. The two-component system is another distinctive function, and it involves the regulation of gene expression in response to environmental signals, such as antibiotic exposure in Gram-negative bacteria (36). However, further explorations are needed to determine whether antibiotic resistance is a trigger of endometriosis. It has been reported that the transcription factor of the endometriosis is involved the regulatory program of endometriosis (37), it can act as a therapeutic target (38). Furthermore, the peptidoglycan biosynthesis function is significantly higher in adenomyosis-endometriosis, and the cell wall of bacteria consists of peptidoglycan and contributes to antibiotic resistance (39). However, there are no reports on the association between peptidoglycan and endometriosis at present.
Conclusions
The present study suggests that the microbiota of the cervix is distinctive in adenomyosis-endometriosis patients, and that Lactobacillus remains dominant. The microbiota in endometriosis and adenomyosis women do not differ in alpha diversity, but the five types of microbiota were distinctive in the genus level, while Atopobium shared the largest proportion in adenomyosis-endometriosis patients. Two-component system, transcription factor, ribosome, peptidoglycan biosynthesis and synthesis and degradation of ketone bodies were possibly the distinctive functions in endometriosis and adenomyosis. These findings may provide a novel view of the microbiota-immune function-endometriosis cross-link system, in which the immune system is associated with the microbiota and endometriosis. However, further studies are needed to understand such network. Furthermore, the present pilot study provides a novel concept that bacteria in lower genital tract may be correlated to the etiology of endometriosis, the bacterial features of endometriosis may become a non-invasive diagnostic method, and even bacterial therapy could be a possible method to treat endometriosis in the future.
Acknowledgments
The authors are very grateful to the Annoroad Gene Technology Co. Ltd (Beijing, China) for their help in 16S ribosomal-RNA gene sequencing and data analysis. The author appreciated the nurses of First Gynecological Ward and Forth Gynecological Ward in Peking Union Medical College Hospital for their co-operation of sample collection before surgery preparation. Jie Guo from CIO Office of Tsing Hua University, conducted the color design of circulized-plot. The author appreciated Ehbio Gene Technology Co. Ltd. (Beijing, China) for providing sequencing analysis methodology and platforms.
Funding: The study was funded by The Ministry of Science and Technology of the People’s Republic of China, National Program on Key Basic Research Project of China (2017YFC1001200).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-1309
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-1309
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-1309). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The Institutional Review Board at the Peking Union Medical College Hospital approved the study (approval No. JS-1532). Informed consent was obtained from all patients, and the study was conducted in accordance with the Declaration of Helsinki Principles (as revised in 2013) and regulations of our institute.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dai Y, Zhou Y, Zhang X, et al. Factors associated with deep infiltrating endometriosis versus ovarian endometrioma in China: a subgroup analysis from the FEELING study. BMC Women’s Health 2018;18:205. [Crossref] [PubMed]
- Ahn SH, Monsanto SP, Miller C, et al. Pathophysiology and Immune Dysfunction in Endometriosis. Biomed Res Int 2015;2015:795976. [Crossref] [PubMed]
- Yuan M, Li D, Zhang Z, et al. Endometriosis induces gut microbiota alterations in mice. Hum Reprod 2018;33:607-16. [Crossref] [PubMed]
- McKinnon BD, Bertschi D, Bersinger NA, et al. Inflammation and nerve fiber interaction in endometriotic pain. Trends Endocrinol Metab 2015;26:1-10. [Crossref] [PubMed]
- Dai Y, Zhang X, Xue M, et al. Not Having Been Breastfed May Protect Chinese Women From Developing Deep Infiltrating Endometriosis: Results From Subgroup Analyses of the FEELING Study. Reprod Sci 2019;26:1158-67. [Crossref] [PubMed]
- Khan KN, Kitajima M, Fujishita A, et al. Pelvic pain in women with ovarian endometrioma is mostly associated with coexisting peritoneal lesions. Hum Reprod 2013;28:109-18. [Crossref] [PubMed]
- Dai Y, Li X, Shi J, et al. A review of the risk factors, genetics and treatment of endometriosis in Chinese women: a comparative update. Reprod Health 2018;15:82. [Crossref] [PubMed]
- Wu J, Xie H, Yao S, et al. Macrophage and nerve interaction in endometriosis. J Neuroinflammation 2017;14:53. [Crossref] [PubMed]
- D'Cruz OJ, Uckun FM. Targeting mast cells in endometriosis with janus kinase 3 inhibitor, JANEX-1. Am J Reprod Immunol 2007;58:75-97. [Crossref] [PubMed]
- Ellis A, Bennett DLH. Neuroinflammation and the generation of neuropathic pain. Brit J Anaesth 2013;111:26-37. [Crossref] [PubMed]
- Murphy K, Mitchell CM. The Interplay of Host Immunity, Environment and the Risk of Bacterial Vaginosis and Associated Reproductive Health Outcomes. J Infect Dis 2016;214 Suppl 1:S29-35. [Crossref] [PubMed]
- Thurman AR, Kimble T, Herold B, et al. Bacterial Vaginosis and Subclinical Markers of Genital Tract Inflammation and Mucosal Immunity. AIDS Res Hum Retroviruses 2015;31:1139-52. [Crossref] [PubMed]
- Mitchell C, Marrazzo J. Bacterial Vaginosis and the Cervicovaginal Immune Response. Am J Reprod Immunol 2014;71:555-63. [Crossref] [PubMed]
- Boskey ER, Cone RA, Whaley KJ, et al. Origins of vaginal acidity: high D/L lactate ratio is consistent with bacteria being the primary source. Hum Reprod 2001;16:1809-13. [Crossref] [PubMed]
- van de Wijgert JHHM, Borgdorff H, Verhelst R, et al. The Vaginal Microbiota: What Have We Learned after a Decade of Molecular Characterization. PLoS One 2014;9:e105998. [Crossref] [PubMed]
- Khan KN, Fujishita A, Hiraki K, et al. Bacterial contamination hypothesis: a new concept in endometriosis. Reprod Med Biol 2018;17:125-33. [Crossref] [PubMed]
- Khan KN, Fujishita A, Masumoto H, et al. Molecular detection of intrauterine microbial colonization in women with endometriosis. Eur J Obstet Gynecol Reprod Biol 2016;199:69-75. [Crossref] [PubMed]
- Chen C, Song X, Wei W, et al. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat Commun 2017;8:875. [Crossref] [PubMed]
- Khan KN, Fujishita A, Kitajima M, et al. Intra-uterine microbial colonization and occurrence of endometritis in women with endometriosis. Hum Reprod 2014;29:2446-56. [Crossref] [PubMed]
- Aagaard K, Ma J, Antony KM, et al. The Placenta Harbors a Unique Microbiome. Sci Transl Med 2014;6:237r-265r.
- Wolfe AJ, Brubaker L. “Sterile Urine” and the Presence of Bacteria. Eur Urol 2015;68:173-4. [Crossref] [PubMed]
- Caporaso JG, Lauber CL, Walters WA, et al. Global patterns of 16s-rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 2011;108 Suppl 1:4516-22. [Crossref] [PubMed]
- Rognes T, Flouri T, Nichols B, et al. VSEARCH: a versatile open source tool for metagenomics. Peer J 2016;4:e2584. [Crossref] [PubMed]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26:2460-1. [Crossref] [PubMed]
- Edgar RC, Flyvbjerg H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 2015;31:3476-82. [Crossref] [PubMed]
- Langille MGI, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16s-rRNA marker gene sequences. Nat Biotechnol 2013;31:814-21. [Crossref] [PubMed]
- Knight R, Vrbanac A, Taylor BC, et al. Best practices for analysing microbiomes. Nat rev Microbiol 2018;16:410-22. [Crossref] [PubMed]
- Takebayashi A, Kimura F, Kishi Y, et al. The association between endometriosis and chronic endometritis. PLos One 2014;9:e88354. [Crossref] [PubMed]
- Baker JM, Chase DM, Herbst-Kralovetz MM. Uterine Microbiota: Residents, Tourists, or Invaders? Front Immunol 2018;9:208. [Crossref] [PubMed]
- Leyendecker G, Bilgicyildirim A, Inacker M, et al. Adenomyosis and endometriosis. Re-visiting their association and further insights into the mechanisms of auto-traumatisation. An MRI study. Arch Gynecol Obstet 2015;291:917-32. [Crossref] [PubMed]
- Polatti F. Bacterial vaginosis, Atopobium vaginae and nifuratel. Curr Clin Pharmacol 2012;7:36-40. [Crossref] [PubMed]
- Mendes-Soares H, Krishnan V, Settles ML, et al. Fine-scale analysis of 16s-rRNA sequences reveals a high level of taxonomic diversity among vaginal Atopobium spp. Pathog Dis 2015;73:ftv020. [Crossref] [PubMed]
- Walther-António MRS, Chen J, Multinu F, et al. Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Med 2016;8:122. [Crossref] [PubMed]
- Mu F, Harris HR, Rich-Edwards JW, et al. A Prospective Study of Inflammatory Markers and Risk of Endometriosis. Am J Epidemiol 2018;187:515-22. [Crossref] [PubMed]
- Chang CY, Lai MT, Chen Y, et al. Up-regulation of ribosome biogenesis by MIR196A2 genetic variation promotes endometriosis development and progression. Oncotarget 2016;7:76713-25. [Crossref] [PubMed]
- Bhagirath AY, Li Y, Patidar R, et al. Two Component Regulatory Systems and Antibiotic Resistance in Gram-Negative Pathogens. Int J Mol Sci 2019;20:1781. [Crossref] [PubMed]
- Yang H, Kang K, Cheng C, et al. Integrative Analysis Reveals Regulatory Programs in Endometriosis. Reprod Sci 2015;22:1060-72. [Crossref] [PubMed]
- Yotova I, Hsu E, Do C, et al. Epigenetic Alterations Affecting Transcription Factors and Signaling Pathways in Stromal Cells of Endometriosis. Plos One 2017;12:e0170859. [Crossref] [PubMed]
- Turner RD, Vollmer W, Foster SJ. Different walls for rods and balls: the diversity of peptidoglycan. Mol Microbiol 2014;91:862-74. [Crossref] [PubMed]


