
Analysis of 2019-nCoV receptor ACE2 expression in different tissues and its significance study
Introduction
The novel coronavirus, now named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), can cause the coronavirus disease 2019 (COVID-19) disease (1). The virus has the characteristics of human-to-human transmission and has a stable transmission capacity. On March 11, 2020, the World Health Organization (WHO) officially announced that COVID-19 had reached global pandemic status. According to the latest statistics released by the world health organization on March 14, 2020, there are currently more than 150,000 patients diagnosed worldwide, of which an estimated 15% are likely to be seriously ill. At present, the most important clinical manifestation of the disease is a pulmonary infection, which is related to the spread of the disease through the respiratory tract. The more severe manifestations are viral pneumonia, bilateral lung infiltration, and respiratory damage (2). Nevertheless, many severe cases are accompanied by further damage to other organs, which cannot be explained simply by the inflammatory storm caused by the novel coronavirus. As a result, the understanding of the disease is flawed in many ways.
It has been reported that the S protein on the surface of the novel coronavirus plays a vital role in the infection and transmission of the virus (3). Further analysis revealed that the receptor to which the S protein binds is angiotensin-converting enzyme 2 (ACE2) (4,5). These findings suggest that binding to the ACE2 receptors is a critical step in the role of the virus and that targeting ACE2 receptor activity may inhibit viral transmission. Some studies have shown that ACE2-expressing cells are also widely present in the intestines, liver, and cardiovascular, and these organs may be as susceptible to infect novel coronavirus as the lungs (6). This study aims to analyze the distribution and expression of ACE2 in different tissues and organs in the body. The results of the study are conducive to the discovery of potential virus-infected organs, and then supply a reference for clinical diagnosis and treatment programs.
Methods
Bioinformatics analysis method
Data sources
Obtain RNAseq transcriptome expression data from the genotype-tissue expression (GTEx) and the Cancer Genome Atlas (TCGA) databases from the University of California Santa Cruz (UCSC) public data source(https://xena.ucsc.edu/). The RNAseq transcriptome was calibrated by Fragments per Kilobase Million (FPKM). The GTEx database contains gene expression and sex data of healthy subjects in different organs. The TCGA database contains gene expression and clinicopathological data of tumor patients in different organs. The tissues and organs included in the database include skin, esophagus, cervix, prostate, breast, bladder, pancreas, thyroid, liver, lung, colon, kidney, and so on.
Data processing and analysis
ACE2 expression data were extracted from all healthy subjects in the GTEx database. This data is classified according to different anatomical sites, and the overall distribution of ACE2 expression in different organs and tissues is analyzed. By comparing the expression of ACE2 in each organ of different sexes, we further explored the correlation between sex and ACE2 expression.
Similarly, we used the TCGA database to analyze the expression distribution of ACE2 in relatively healthy tissue around the cancer of different organs. We obtained ACE2 expression data for different tissues and plotted histograms for overall observation. We also classified the samples according to sex and discussed the correlation between sex and ACE2 expression in tumor tissues.
We further combined all samples and analyzed the expression of ACE2 in different tissues, as well as the associations between the sexes. The analyses of the different groups validate each other.
Statistical methods
Statistical analysis of all RNA-seq transcriptome data was conducted using R v3.4.1(https://www.r-project.org/). The Games-Howell Test was used to compare the differences in ACE2 expression between pairs of different organs, and the t-test was used to compare the differences in ACE2 expression in different organs of different sexes. P<0.05 was considered statistically significant.
Results
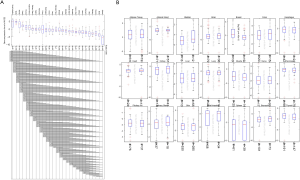
Based on bioinformatics technology, we collected ACE2 gene expression data of 6,738 healthy subjects in different organs and sex information of different samples in the GTEx database. According to the organs to which the samples belong as a group, we plotted the histograms of ACE2 expression values in different organ groups. The results suggest that there are differences in ACE2 genes for different organs; the highest expression of ACE2 is in the small intestine, and spleen has the lowest expression (Figure 1A). Further combining sex factors, we found that male and female ACE2 expression levels in the same organ were not significantly different (Figure 1B).

The TCGA database extracted ACE2 expression data from 618 different tissues. The results are consistent with the analysis of the GTEx database. The kidney has the highest expression of ACE2, the second one is colon tissue, and the lowest expression of ACE2 is in skin tissue (Figure 2A). Similarly, there is not much difference in ACE2 expression between men and women in the same organ (Figure 2B).
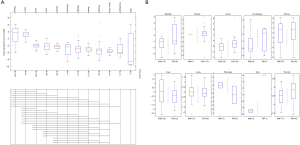
Finally, we combined the GTEx and TCGA database samples with a total of 7,356 cases. We compared the expression levels of ACE2 in different organs and found that the expression was highest in the small intestine. The expression in testis, kidney, heart, thyroid, adipose tissue, salivary glands, and other tissues was also at a high level, while the spleen, brain, muscle, pituitary, and skin tissue ACE2 expression was low (Figure 3A). The effect of sex on the expression of ACE2 in different organs was not significant (Figure 3B).
Discussion
ACE2 is the first human angiotensin-converting enzyme homolog discovered in 2000, encoding a gene on the X chromosome (7,8). ACE2 is a zinc metalloproteinase, a type I transmembrane protein. The structure of ACE2 includes a signal peptide, a transmembrane domain, and a metalloproteinase active site having a HEXXH zinc-binding domain. It can degrade Ang I to produce the non-peptide Ang 1-9, and degrade Ang II to produce the heptapeptide Ang 1-7 (9,10). ACE2 was first found to be expressed in the heart, kidney, and testis. Later studies confirmed that ACE2 is also widely expressed in the lung, brain, and digestive tract (11). For example, ACE2 is mainly distributed in type II alveolar cells (AT2 cells) in the lung tissue and is also distributed in small amounts in type I alveolar cells (AT1 cells), airway epithelial cells, fibroblasts, endothelial cells and macrophages (12).
2019-nCoV rely on the S protein on the surface of the virus to bind to ACE2 on the surface of tissue cells to invade the body, and the trimeric structure of 2019-nCoV S protein is more likely to bind to the ACE2 protein on the cell surface (4). Therefore, the affinity of 2019-nCoV is 10 to 20 times higher than of the severe acute respiratory syndrome (SARS) virus, which is also a coronavirus (13-15). The high affinity is why 2019-nCoV is highly contagious and also suggests that our cells and tissues that highly express ACE2 may be the main target of SARS-CoV-2, and the expression level of ACE2 may be of great value in predicting the invasion of tissues by 2019-nCoV.
Based on bioinformatics technology, we have conducted in-depth mining of the GTEX database and TCGA database. The aim is to explore the expression level of ACE2 in different tissues, to provide a reference for clinical progress, diagnosis and treatment plan formulation, and pretreatment for detecting potential infection of organs in severe patients. For example, if the is kidney-highly expressed, then ACE2 is a potential target for 2019-nCoV. Therefore, the treatment of severe patients can increase the prevention of renal failure and renal replacement therapy.
With the increasing number of 2019-nCoV studies, some clinicopathological evidence also supports our analysis. The recent literature reports that 2019-nCoV has been detected in the gastrointestinal tract, saliva, and urine of pneumonitis patients infected by 2019-nCoV (6). And other studies have shown that there are also ACE2-rich cells in the intestine, liver, and cardiovascular (11,16,17). These organs may also be infected with 2019-nCoV, like the lungs. National Institutes of Health (NIH) study shows that 2019-nCoV may attack testes, and the targets are mainly concentrated in spermatogonia, mesenchymal cells, and support cells in the testis (18). The Journal of Forensic Medicine reported Professor Liu Liang’s general anatomy of a patient who died of 2019-nCoV pneumonia infection within 15 days. The results showed that the lung was significantly injured, the epicardium was slightly edema, the myocardial section was gray-red fish-like, the small intestine was dilated and narrowed, and the kidney was granular and contracted. At the same time, pathological reports suggest that 2019-nCoV plays a role in pathological changes of multiple organs such as the lung, spleen, hilar lymph nodes, heart and blood vessels, liver, and gallbladder, and kidneys. However, considering that an autopsy is performed on a case-by-case basis, whether the above changes are related to viral infection-related damage remains to be further studied (19). Whether 2019-nCoV can cause explosive myocarditis is inconclusive, but some epidemiological investigations have shown that the death of some severe patients may be related to explosive myocarditis, but this needs to be further determined (20). For adipose tissue that may overexpress ACE2, Professor Lu of Peking University has reported that it may also be related to the susceptibility of obese patients to infection with 2019-nCoV (21). In summary, these studies are consistent with our analysis results. Our results may provide some theoretical basis for clinical data or observations and provide references for in-depth research on the invasion of 2019-nCoV to the body and various tissues.
Conclusions
Given the high affinity of the new crown virus and the ACE2 receptor, tissues with high expression of ACE2 will be more likely to be the target of the attack of the new crown virus. This study used bioinformatics to analyze the expression level of ACE2 in healthy tissues of normal subjects in the GTEX database, and also analyzed the expression level of ACE2 in relatively healthy tissue around the different organs cancer in the TCGA database. Finally, the two databases were combined to analyze again, and the conclusions obtained were highly consistent. The expression of ACE2 in different tissues is significantly different, and this expression difference is not affected by sex. The expression of ACE2 is higher in the small intestine, testis, kidney, heart, thyroid, adipose tissue, salivary gland, and other tissues, and the results of related clinical reports are consistent with our analysis. This study clarified the distribution and expression of ACE2 in various tissues and organs of the human body. The results of this study are conducive to the discovery of potential infection target organs of the virus and provide a reference for the formulation of diagnosis and treatment plans for patients with future clinical novel coronavirus infections and the pretreatment of potentially infected organs in severe patients.
Acknowledgments
Thanks for Jianing Xi’s selfless help and guidance in Bioinformation Analysis.
Funding: Jieping Wu Medical Foundation supported this study (No. 320.6750.19021) and the 2018 China Medical University Young Backbone Support Program (No. QGZ2018011).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-4281). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lai CC, Shih TP, Ko WC, et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int J Antimicrob Agents 2020;55:105924. [Crossref] [PubMed]
- Guo YR, Cao QD, Hong ZS, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res 2020;7:11. [PubMed]
- Yan R, Zhang Y, Li Y, et al. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science 2020;367:1444-8. [Crossref] [PubMed]
- Zhang H, Penninger JM, Li Y, et al. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med 2020;46:586-90. [Crossref] [PubMed]
- Batlle D, Wysocki J, Satchell K. Soluble angiotensin-converting enzyme 2: a potential approach for coronavirus infection therapy. Clin Sci (Lond) 2020;134:543-5. [Crossref] [PubMed]
- Guan W, Ni Z, Hu Y, et al. Clinical characteristics of 2019 novel coronavirus infection in China. MedRxiv 2020. doi: https://doi.org/ [Crossref]
- Santos RA, Sampaio WO, Alzamora AC, et al. The ACE2/Angiotensin-(1-7)/MAS Axis of the Renin-Angiotensin System: Focus on Angiotensin-(1-7). Physiol Rev 2018;98:505-53. [Crossref] [PubMed]
- Ortiz-Melo DI, Gurley SB. Angiotensin converting enzyme 2 and the kidney. Curr Opin Nephrol Hypertens 2016;25:59-66. [Crossref] [PubMed]
- Turner AJ, Tipnis SR, Guy JL, et al. ACEH/ACE2 is a novel mammalian metallocarboxypeptidase and a homologue of angiotensin-converting enzyme insensitive to ACE inhibitors. Can J Physiol Pharmacol 2002;80:346-53. [Crossref] [PubMed]
- Qin S, Zhou YJ, Liu Y, et al. (Expression and significance of ACE2-Ang-(1-7)-Mas axis in the endometrium of patients with polycystic ovary syndrome). Zhonghua Yi Xue Za Zhi 2013;93:1989-92. [PubMed]
- Hamming I, Timens W, Bulthuis ML, et al. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol 2004;203:631-7. [Crossref] [PubMed]
- Zhu L, Carretero OA, Xu J, et al. Activation of angiotensin II type 2 receptor suppresses TNF-α-induced ICAM-1 via NF-кB: possible role of ACE2. Am J Physiol Heart Circ Physiol 2015;309:827-34. [Crossref]
- Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020;367:1260-3. [Crossref] [PubMed]
- Xu X, Chen P, Wang J, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci 2020;63:457-60. [Crossref] [PubMed]
- Zhou P, Yang XL, Wang XG, et al. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. BioRxiv 2020. doi: https://doi.org/ [Crossref]
- Vuille-dit-Bille RN, Camargo SM, Emmenegger L, et al. Human intestine luminal ACE2 and amino acid transporter expression increased by ACE-inhibitors. Amino Acids 2015;47:693-705. [Crossref] [PubMed]
- Wu HT, Chuang Y, Huang C, et al. Loss of angiotensin converting enzyme II (ACE2) accelerates the development of liver injury induced by thioacetamide. Exp Anim 2018;67:41-9. [Crossref] [PubMed]
- Wang Z, Xu X. scRNA-seq Profiling of Human Testes Reveals the Presence of ACE2 Receptor, a Target for SARS-CoV-2 Infection, in Spermatogonia, Leydig and Sertoli Cells. Preprints 2020. doi: . [Crossref]
- liu Q, Wang R, Qu G, et al. Gross examination report of a COVID-19?death autopsy. Fa Yi Xue Za Zhi 2020;36:19-21.
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497-506. [Crossref] [PubMed]
- Jia X, Yin C, Lu S, et al. Two Things about COVID-19 Might Need Attention. Preprints 2020. doi: . [Crossref]


