
Expression of p42.3 in non-small cell lung cancer
Introduction
Lung cancer has historically been the leading cause of cancer morbidity and mortality in the world (1), while non-small cell lung cancer (NSCLC) accounts for about 85% of primary lung cancers (2).
The p42.3 gene (GenBank, DQ150361) is a highly conserved mammalian gene with a cDNA length of about 4.0 kb. It is highly homologous to the c9orf140 gene located on the human chromosome 9q34.3 and encodes a protein composed of 389 amino acids with a molecular weight of 42.3 kDa; therefore, it has been named the p42.3 gene. p42.3 is specifically expressed in a variety of tumor cell lines. The expression of p42.3 is cell cycle-dependent, and its mRNA expression in G1 and M phases are higher than that in S and G2 phases examined by RT-PCR. Among them, the M phase has the highest expression and gradually decreases after cell division, indicating that this gene may be involved in cell cycle regulation. This gene has a regulatory effect on the key proteins involved in cell cycle regulation, including the CHK2 and cyclin B1 proteins of gastric cancer (GC) cell lines, suggesting that it may be involved in tumor development as an oncogene (3-5).
Furthermore, the overexpression of p42.3 has been proven to be closely related to the clinical stage of malignant melanoma, the 5-year survival rate of colorectal cancer (CRC) patients, and the histological grade of glioma (6-8). However, little is known about the relationship between p42.3 and NSCLC. Therefore, in this study, we explored the expression of p42.3 in NSCLC by bioinformatics analysis, and discussed the relationship between p42.3 protein expression and clinicopathological characteristics in combination with clinical cases.
Methods
Bioinformatic analysis using FireBrowse
The analysis of the p42.3 expression in the solid tumors and corresponding normal tissues was performed with data from The Cancer Genome Atlas (TCGA). The data generated by the TCGA was analyzed using FireBrowse (http://firebrowse.org/) (9).
Bioinformatic analysis using UCSC Xena browser
The level 3 data of patients with primary NSCLC in TCGA-NSCLC (TCGA data on NSCLC) were extracted using the UCSC Xena browser (https://xenabrowser.net/). The p42.3 mRNA expression and DNA methylation in patients with primary lung adenocarcinoma (LUAD) or lung squamous cell carcinoma (LUSC) were examined using data from the TCGA-LUAD (TCGA data on LUAD) and TCGA-LUSC with the UCSC Xena browser. Kaplan-Meier curves for the overall survival (OS) rates after initial therapy were also generated using the same browser.
Bioinformatic analysis using cBioPortal for cancer genomics
p42.3 genetic alterations from the TCGA-LUAD and TCGA-LUSC were examined using the cBioPortal for Cancer Genomics (http://www.cbioportal.org/) website (10,11). Only pathways with a P value <0.05 were included.
Patients and samples
NSCLC patients (n=142, 72 LUSC patients and 70 LUAD patients) who were diagnosed and underwent surgery at Beijing Hospital between 2005 and 2009 were included. None of these patients received anti-tumor therapy before surgery. The tumor tissue and paired normal tissue paraffin specimens were assigned to the tumor group and the control group, respectively. These tissue samples were processed in tissue microarrays (TMA). The clinicopathological data of the patients were collected and collated. This study is a retrospective observational study. Only paraffin specimens from tumor tissues of patients with NSCLC that were previously preserved by the subjects were collected for the study. No intervention measures were taken for the subjects. The collection of information and the publication of research results do not contain unique information that can identify the subject. Informed consent is not required. The trial was conducted in accordance with the Declaration of Helsinki. The study was approved by the Ethics Review Committee of Beijing Hospital (No. 2020BJYYEC-111-01).
Immunohistochemistry (IHC)
The TMA paraffin section (4 µm thick) was baked at 65 °C for 60 minutes. Next, it was dewaxed in xylene, and hydrated with graded alcohol water and a phosphate-buffered saline (PBS, pH 7.2–7.4). Treatment with a 3% hydrogen peroxide solution for 10 minutes was applied to block endogenous peroxidase activity. Following these steps, we heated the TMA in a 1X citrate buffer (pH 6.0) for 10 minutes in the microwave, and natural cooling restored the protein to its original spatial conformation. Goat serum (SP KIT-B2) was used to reduce nonspecific binding. Then, the SAPCD2 polyclonal antibody (PA5-60632) (diluted 1:3,000) was selected as the p42.3 protein antibody, and the TMA was incubated in SAPCD2 overnight at 4 °C. After incubating with immunochromogenic reagent (Kit-5020) at room temperature for 20 minutes, we added 3,3'-diaminobenzidine (DAB) for 2 minutes. Then hematoxylin redyeing was completed with 1% hydrochloric acid ethanol differentiation, 1% ammonia regain blue, and gradient alcohol dehydration.
Following this, the histopathological evaluations were performed independently by 2 pathologists regardless of the patient’s clinical data. The positive expression of p42.3 protein showed brownish yellow granules in the cytoplasm. The positive staining area ≥1% was defined as positive. The qualitative score of immune response was given according to the microscopic staining intensity in the following manner: staining intensity (–) = negative staining, no coloration of tumor cytoplasm; (+) = weak-positive staining, sparse brownish yellow granules observed in the cytoplasm of tumor cells; (++) = medium staining, the staining strength was between weak-positive and strong-positive; (+++) = strong-positive staining, deeply stained brownish yellow granules observed in the cytoplasm of tumor cells (Figure 1).

Histological grading method
There is no recognized, specific grading system for most lung cancers (12). The hematoxylin and eosin (HE) staining results were scored according to the tumor cell differentiation degree reference for the current clinical general grading method in which a higher level of differentiation indicates that the tumor cells are closer to the normal source tissues. The grading is as follows: high differentiation (grade I), medium differentiation (grade II), and low differentiation (grade III).
Statistical analysis
The statistical analyses were performed using SPSS 22.0 (SPSS, Chicago, IL, USA). Pearson’s chi-square (χ2) test was used to compare the differences in p42.3 protein expressions between the NSCLC tumor tissues and the adjacent normal tissues, and between the LUSC and LUAD tumor tissues. The relationship between age and positive p42.3 protein expression was also evaluated using the χ2 test. Next, the association between the p42.3 protein expression and the clinicopathological features were evaluated using Fisher’s exact test. In all cases, P values <0.05 were considered to be statistically significant.
Results
Part 1: Bioinformatics analysis
p42.3 was significantly upregulated in both LUAD and LUSC compared with the normal lung tissues
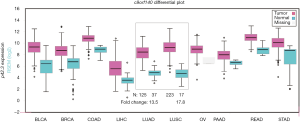
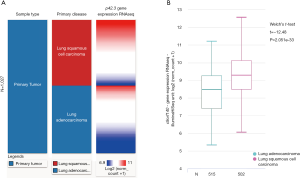
We characterized p42.3 mRNA expression in several types of solid tumors, including LUAD and LUSC, using FireBrowse for data mining. Our results indicated that p42.3 expression in the LUAD tissues was approximately 13.5-fold higher than in the normal lung tissues. Furthermore, the LUSC tissues were about 17.8-fold higher than the normal lung tissues (Figure 2). For further comparisons, the p42.3 mRNA RNA-sequencing data extracted from TCGA-LUAD and TCGA-LUSC were analyzed. Heatmaps and subsequent comparisons showed that the p42.3 expression in the LUSC tissues was significantly higher than that in the LUAD tissues (Figure 3A,B).
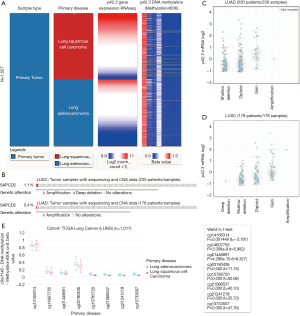
LUSC had a lower level of p42.3 DNA methylation and a higher level of p42.3 DNA amplification than LUAD
We explored the potential mechanisms of p42.3 expression dysregulation and observed that the level of p42.3 DNA methylation was significantly lower in the LUSC patients than in the LUAD patients after comparing p42.3 expression and its DNA methylation data (Figure 4A). Afterward, we examined copy number alterations (CNAs) from the TCGA-LUAD and TCGA-LUSC data. The p42.3 mutation was observed in 0.4% of LUAD and 1.1% of LUSC cases (Figure 4B). Amplification was the predominant type of mutation, and it was associated with an increase of p42.3 mRNA expression in LUAD and LUSC (Figure 4C,D). In addition, we visualized methylation results in Figure 4E.
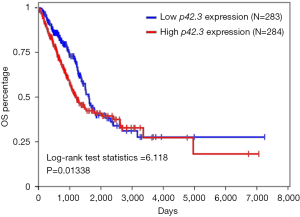
p42.3 DNA mutation was not significantly associated with worse OS in LUAD (lack of survival data in LUSC)
We investigated the associations between p42.3 DNA mutation and the survival rates in LUAD and LUSC patients. The survival curve failed to indicate that LUAD patients with a p42.3 amplification were associated with a worse OS rate (Figure 5). On the other hand, although p42.3 amplification was more significant in LUSC patients, there was no relevant survival information in TCGA.
Part 2: Real-world research
p42.3 protein expression in NSCLC tumor tissues was significantly higher than in adjacent normal tissues
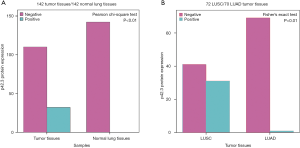
A total of 142 NSCLC patients were included in the IHC evaluation (tissue separation from the slides were excluded), which included 72 LUSC and 70 LUAD samples. IHC staining showed that p42.3 protein was mainly expressed in the cytoplasm of NSCLC with variable intensity (Figure 1). Furthermore, p42.3 protein expression in NSCLC tumor tissues was significantly higher than that in normal tissues (32/142, 22.5% vs. 0/142, 0%; P<0.01) (Figure 6A).
p42.3 protein expression in LUSC was significantly higher than LUAD
Then, tumor tissue samples were further divided into the LUSC and LUAD groups according to the pathological classification. According to the results of the Fisher’s exact test, the p42.3 protein expression in the LUSC was significantly higher than that in the LUAD group (31/72, 43.1%, vs. 1/70, 1.4%; P<0.01) (Figure 6B). The clinicopathological features of LUSC patients in the real world were further analyzed to explore the significance of p42.3 upregulation.
Association of p42.3 protein expression with clinicopathological features in LUSC
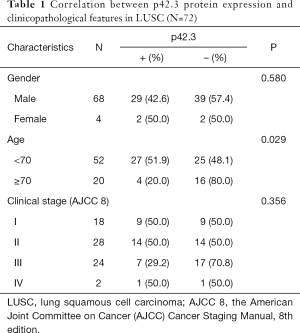
The clinical and pathological information of 72 LUSC patients, including 68 males (94.4%, 68/72) and 4 females (5.6%, 4/72), were analyzed. At the time of surgery, the ages of the patients ranged from 31 to 77 (median age 64.4) years old. All tumors stage classifications were conducted according to the American Joint Committee on Cancer (AJCC) Cancer Staging Manual, 8th edition (stage I–IV; no carcinoma in situ) (Table 1). No significant correlation was found between p42.3 protein expression and gender (Fisher, P=0.58). The positive expression rate of p42.3 protein was significantly different between patients younger than 70 years and those 70 years or older (51.9%, 27/52 vs. 20%, 4/20; P=0.029). Due to the small sample size of stage IV cases, which might have caused statistical bias, Fisher’s exact test was further used to analyze the correlations between clinical stage or histological grade and p42.3 protein expression. No significant difference was found between different clinical stages (P=0.356). However, for the patients with advanced histological grade, the IHC staining was stronger, which indicated that high p42.3 protein expression was correlated with lower differentiation (P=0.043) (Table 2).
Full table

Full table
Discussion
Based on the current data from TCGA-LUAD and TCGA-LUSC databases, the abnormal expression of p42.3 in LUAD and LUSC tissues has been observed. Furthermore, we found that p42.3 expression was significantly higher in LUSC than in LUAD. Approximately 1.1% of LUSC cases had p42.3 amplification, compared with only 0.4% for LUAD. We also observed that some CpG loci of the p42.3 gene had a higher methylation level in LUAD than in LUSC, which suggests that epigenetic changes are important mechanisms for p42.3 dysregulation in NSCLC. These findings help to explain why p42.3 expression is significantly higher in LUSC than in LUAD. Most importantly, we observed that high p42.3 protein expression correlated with worse differentiation, suggesting that p42.3 may have an important prognostic value in LUSC.
As an oncogene, p42.3 upregulation is also prognostic in some cancers. In the GC cell line (BGC823), p42.3 was found to participate in malignant transformations, while the silencing of p42.3 expression could significantly inhibit the proliferation and oncogenicity of tumor cells (3). Studies on CRC showed that the expression of p42.3 was an independent prognostic factor in CRC patients (P=0.030). Patients with high expression of p42.3 had a poor prognosis (P=0.033) (7). Furthermore, high p42.3 expression in patients with primary hepatocellular carcinoma (HCC) was significantly correlated with worse differentiation (P=0.031) (13). Besides, high expression of p42.3 was correlated with the clinical stage of melanoma patients (P=0.045) (6) and high histological grade of glioma (P<0.01) (8).
In regards to the driving mechanisms of p42.3 expression in tumorigenesis, recent studies have shown that the p42.3 gene might be a regulatory factor involved in the signaling pathways. It was found that in GC cell lines, p42.3 protein expression was negatively correlated with the expression of microRNA-29a (miR-29a), which directly targeted p42.3 3'UTR, while the knockout of the p42.3 gene could inhibit tumor cell proliferation and induce cell cycle arrest (14). Meanwhile, miR-29a was shown to significantly inhibit the proliferation and invasion of the GC cell lines, while the expression of miR-29a in human GC lines was significantly downregulated (15).
These findings are consistent with the observations for GC, HCC, and other tumor cell lines: the silencing of the p42.3 gene leads to a down-regulation of the cell cycle regulatory protein cyclinB1 and an up-regulation of CHK2, inducing changes in the biological processes of proliferation, invasiveness, and malignant transformation (3,13,16). The decreased expression rate of p42.3 protein in LUSC patients aged 70 years or older is consistent with the decreased metabolic rates in elderly patients. Therefore, it is speculated that the p42.3 protein may be a significant regulatory factor involved in this signaling pathway. It is of great significance to further study the role of p42.3 in NSCLC.
In conclusion, the present study confirmed that both genetic and epigenetic alterations contribute to the dysregulation of p42.3 in NSCLC. The high expression of p42.3 protein was an independent factor of a worse pathological differentiation in LUSC. Therefore, there exists a potential for p42.3 to be developed as a cost-effective biomarker of LUSC prognosis.
To further explore the biological functions, prognostic value, and potential mechanisms of p42.3 in NSCLC, prospective studies with expanded sample sizes should be conducted. Meanwhile, findings from cell transfection, and cell proliferation, scratch wound healing, and migration and invasion assays will continue to be reported.
Acknowledgments
We would like to thank the State Key Laboratory of Molecular Oncology, Department of Etiology and Carcinogenesis, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College for providing the laboratory.
Funding: This work was supported by the CAMS Innovation Fund for Medical Sciences (2018-I2M-1-002) and Beijing Hospital Research Fund for Clinical medicine (BJ-2018-016).
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-2928
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2928). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Informed consent is not required. This study is a retrospective observational study. Only paraffin specimens from tumor tissues of patients with NSCLC that were previously preserved by the subjects were collected for the study. No intervention measures were taken for the subjects. The collection of information and the publication of research results do not contain unique information that can identify the subject. The trial was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Review Committee of Beijing Hospital (No. 2020BJYYEC-111-01). Individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Bender E. Epidemiology: the dominant malignancy. Nature 2014;513:S2-3. [Crossref] [PubMed]
- Xu X, Li W, Fan X, et al. Identification and characterization of a novel p42.3 gene as tumor-specific and mitosis phase-dependent expression in gastric cancer. Oncogene 2007;26:7371-9. [Crossref] [PubMed]
- Hartwell LH, Kastan MB. Cell cycle control and cancer. Science 1994;266:1821-8. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Liu H, Zhu M, Li Z, et al. Depletion of p42.3 gene inhibits proliferation and invasion in melanoma cells. J Cancer Res Clin Oncol 2017;143:639-48. [Crossref] [PubMed]
- Yuan XS, Zhang Y, Guan XY, et al. p42.3: a promising biomarker for the progression and prognosis of human colorectal cancer. J Cancer Res Clin Oncol 2013;139:1211-20. [Crossref] [PubMed]
- Wan W, Xu X, Jia G, et al. Differential expression of p42.3 in low- and high-grade gliomas. World J Surg Oncol 2014;12:185. [Crossref] [PubMed]
- Zhou S, Wang P, Su X, et al. High ECT2 expression is an independent prognostic factor for poor overall survival and recurrence-free survival in non-small cell lung adenocarcinoma. PLoS One 2017;12:e0187356. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Yang X, Lin D. Changes of 2015 WHO histological classification of lung cancer and the clinical significance. Zhongguo Fei Ai Za Zhi 2016;19:332-6. [PubMed]
- Sun W, Dong WW, Mao LL, et al. Overexpression of p42.3 promotes cell growth and tumorigenicity in hepatocellular carcinoma. World J Gastroenterol 2013;19:2913-20. [Crossref] [PubMed]
- Cui Y, Su WY, Xing J, et al. MiR-29a inhibits cell proliferation and induces cell cycle arrest through the downregulation of p42.3 in human gastric cancer. PLoS One 2011;6:e25872. [Crossref] [PubMed]
- Chen L, Xiao H, Wang ZH, et al. miR-29a suppresses growth and invasion of gastric cancer cells in vitro by targeting VEGF-A. BMB Rep 2014;47:39-44. [Crossref] [PubMed]
- Mao L, Sun W, Li W, et al. Cell cycle-dependent expression of p42.3 promotes mitotic progression in malignant transformed cells. Mol Carcinog 2014;53:337-48. [Crossref] [PubMed]


