
Human papilloma virus (HPV) profiles in breast cancer: future management
Introduction
Globally, breast cancer (BC) is the most common cancer among the women registering a total of 2.08 million new cases (11.6% of all new cases among females) in the year 2018 alone (1). Accounting for 15% of the total cancer-related deaths, it is the first most common cause of cancer deaths among women, worldwide (1). In Indian context, BC remains the most frequent (27.7%) cancer among women with the urban and metropolitan regions reporting high rates of incidence than rural region (1,2). Going by the numbers, in 2018 about 87,090 women died due to BC in India (11.1% of total women cancer) (1).
The BC has several etiological factors like prolonged or elevated exposure to estrogen due to early age of menarche (younger than 12 years), nulliparity, late age of menopause (over 55 years), exposure to high doses of ionizing radiation, regular alcohol consumption and high fat diet (3). Among the different etiological factors, infection with several viruses has also been reported in BC (4). However, these etiological factors were involved in only 20–50% of BC cases (5). Recently, different studies suggested association of human papillomavirus (HPV) with BC (6). But, frequency of HPV infection in BC varied widely (1.6–86%) among different studies (7,8). Inconsistent HPV infection was also reported in different molecular subtypes of BC (9,10). The possible mode of HPV transmission in breast and its role in breast carcinogenesis are not well studied. In this review our aim is to discuss the role of HPV infection in breast carcinogenesis and its future management.
Association of HPV infection with BC
Prevalence of HPV infection in breast
Recently, HPV infection in BC in different population around the world was reported by several authors (Table 1). However, many of them have not identified any HPV DNA in breast tumour. The prevalence of HPV in BC varied widely from 1.6–86.2% among the different continents of the world (7,8). According to screening methods, comparatively high frequency of HPV was detected in polymerase chain reaction (PCR) with sequencing or in-situ hybridization than only PCR method alone (Figure 1A). While a comparatively lower frequency of HPV DNA was found when the tissue source was formalin fixed paraffin-embedded tissue (PET) than the cryo-preserved tissue (CPT), the reason can be attributed towards the fact that the total DNA is severely degraded during the whole process of formalin fixation and paraffin embedding (47). So, this detection based difference in results might account partly for the wide range of frequency of HPV infection in BC, as reported by several studies (Figure 1B). On the other hand, HPV infection did not show significant variation among the different continents of the world (Figure 1C). To date, nine HPV types (HPV6, 11, 16, 18, 31, 33, 35, 45 and 52) are evident in BC across different population of the world. The prevalence of these HPV types showed variation among different population. The HPV16 was prevalent in American BC patients, whereas HPV18 and HPV33 were frequent in Australian and Chinese BC patients (Table 1). Apart from the above mentioned three subtypes, prevalence of other subtypes in BC patients among different population are as follows: HPV6/HPV11 in 5–12.6% patients of Iran and Spain (39,40), HPV31 in 1.5–11.5% patients of Brazil and UK (37,48), HPV35 in 16–19.2% of patients of Thailand and UK (37,49), HPV45 in 23% of UK BC patients (37) and HPV52 in 1.5–11% of Brazil, UK and Thailand patients (37,48,49).
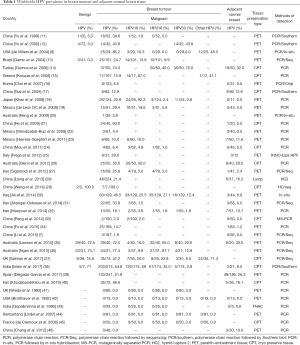
Full table
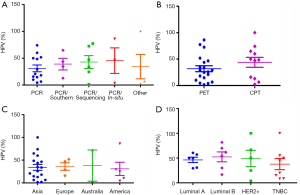
HPV infection was also evident among the different subtypes of BC (Table 2). Among these subtypes, comparative high HPV infection was observed in Luminal B than other BC subtypes indicating that these cells might be favourable for HPV survival or may serve as an initial target of HPV infection due to the cooperative interaction with HER2 as well as ER (Figure 1D) (55,56). HPV infection in Triple Negative Breast Cancer (TNBC) varied from 15–50% in different studies, in which HPV16 was the most prevalent subtype (Table 2). In addition, HPV infection was also reported in adjacent normal and benign breast tissue (Table 1) (57) as well as in BC cell lines MDA-MB-175-VII, SK-BR-3 and MCF7 (20,38). HPV infection was also reported in nipple tissue, breast ductal lavage, nipple discharge and even from breast milk (8,58-62). Interestingly, presence of HPV was also observed in the serum-derived extracellular vesicles (58). In many studies, the presence of HPV genome in Indian, Italian and Australian BC patients was confirmed by sequencing analysis apart from PCR based methods (35,38,58).
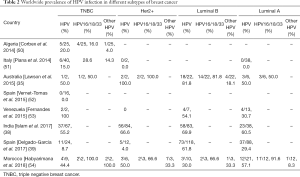
Full table
Significant association between HPV infection, clinical grade, young age of the patients and histology were reported by different investigators worldwide (38,53,56), which further establish the clinical implication of HPV infection in BC. In addition, HPV associated poor prognosis of BC patients was also reported by our group and Ohba et al. (38,56).
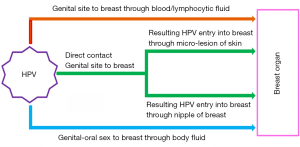
Possible route of HPV infection in breast:
HPV infection can be transmitted through both sexual and nonsexual contacts. The genital HPV is mostly transmitted by direct skin-to-skin contact during sexual intercourse with an infected person (63). Generally, HPVs enter into the body through the skin and epidermal injuries, mucous membranes, skin abrasions and infects the cells of the basal layer of the stratified epithelium (64). The internalization of virions occurs slowly by endocytosis of clathrin coated vesicles in the presence of heparin sulphate. This ultimately leads to the transport of viral DNA to the nucleus and in the process disruption of the intracapsomeric disulphide bonds of the viral capsid occurs in the reducing environment of the cell (65-70). However, there can be three possible mode of HPV infection in breast tissue (Figure 2). According to the first one, HPV may be transmitted to breast from the genital region of the patients having a previous history of HPV-positive uterine cervical cancer (CACX) through blood, lymphatic systems or any other body fluid (71). It may be the case where a secondary malignant transformation of breast tissue could occur by an HPV infected malignant cell, which is derived from the primary tumour of any other site (72,73). It may also be due to spill over of HPV virion to the circulation system from HPV infected primary tumour site (74). As per the second mechanism, transmission of HPV can occur to breast from any oral site due to oral sexual practices (46). Third one suggests that the transmission of HPV may occur to breast by nipple or micro-lesion of breast skin due to genital-breast sexual activity (75,76).
Molecular profiles of HPV in BC
The persistent high-risk (hr) HPV infection are well known prerequisite factor for clinical progression and the development of Cervical intraepithelial neoplasia III (CIN III) and CACX (77-79). The persistent infections with hrHPVs have been identified as an essential but not sufficient factor in the pathogenesis of anogenital and other epithelial carcinomas (80). It was evident that sequential changes in the molecular profiles (genetic/epigenetic expression) of HPV occurred during development of tumour. Recent studies have shown that the majority (86–100%) of HPV genome present in breast tissue in an integrated form, an important step of HPV induced normal epithelial cell transformation as well as carcinogenesis (Table 3) (85). On the other hand, low copy number of HPV genome with range 0.00054–9.3 copies/cell in breast tumor was reported by different investigators including our group (Table 3). Based on sequence variation of the HPV genome, four naturally occurring lineages have been characterized like European-Asian (A), African-1(Af-1) (B) African-2(Af-2) (C) and Asian-American-North American (D) (86). Among these, American-North American (D) lineage was associated with the virulence property (87). Our previous sequence variation analysis of E6-E7 and LCR regions of HPV16 genome revealed that “A” lineage was frequent in BC (64.2%, 36/56) followed by D (33.9%) and B (1.78%) (38). Among these, frequent variants such as 7521 G > A at LCR and 350T > G at E6 regions indicated their importance in the process of carcinogenesis (88). HPV genome is functionally subdivided into three regions: early, late and the regulatory-long control region (LCR) or non-coding region (NCR), each are separated by two polyadenylation (pA) sites: early pA (pAE) and late pA (pAL) sites (Figure 3) (89). After HPV infection and capsid uncoating, P97 promoter derived early poly-cistronic mRNA transcript is responsible for production of early response proteins i.e., E1, E2, E4, E5, E6 and E7 by differential splicing (90). On the other hand, the poly-cistronic mRNA transcript from the late promoter P670 through differential splicing could produce E1, E2, E4, L1 and L2 proteins. Our previous study showed high methylation in p97 promoter (97%) and enhancer (51%) at LCR region of HPV16 genome, indicating the importance of this epigenetic modification in regulation of the viral genome expression (38) (Table 3).
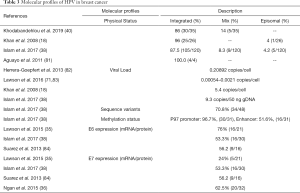
Full table
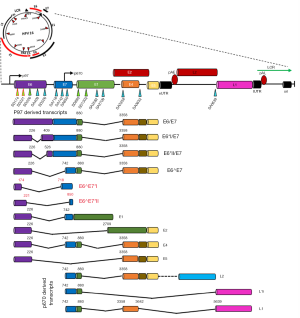
The expression of E6 and E7 oncogenes have their significant biological implications in HPV induced carcinogenesis. The E6/E7 transcripts were detected in 24–100% of BC samples by different researchers including our group (Table 3). Apart from the existing transcripts of E6/E7, two novel fusion transcripts of E6/E7 (E6^E7*I, E6^E7*II) in breast tumour were detected by us suggesting the underlying differences in molecular pathogenesis of HPV in BC compared to other cancers (Figure 3) (38). Going further, different investigators including our group detected the E6/E7 protein expression in 24–76% breast samples indicating functional relevance of HPV in breast tumour tissue (Table 3) (35). In addition, E6 and E7 expression was also evident in adjacent normal tissue, nipple tissue and epithelial layer of normal breast skin (8,38,71).
Molecular pathogenesis of HPV associated BC
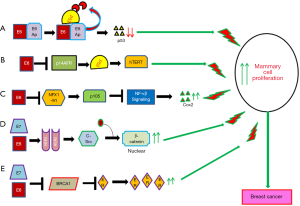
The molecular mechanism of HPV infection in promoting cervical cancer development and progression has been studied comprehensively (91). However, the exact mechanism by which HPV induces or promotes breast carcinogenesis is not well defined yet. It was evident that the E6 and E7 oncoproteins of HPV16 could immortalize human mammary epithelial cells through inactivation of p53 and RB respectively indicating their importance in cellular transformation (55,92). Different in-vitro studies showed association of E6/E7 with multiple cellular pathways in transformation of mammary epithelial cells (Figure 4) (5). Among these pathways, E6/E7 could down regulate P53, NFX1 and BRCA1 resulting up regulation of CoX2, NF-κβ and ER associated pathways (72,93-97). On the other hand, E6/E7 could stabilize HER2 receptor resulting in the activation of beta-catenin and thus enhance cellular proliferation (Figure 3) (55,98). Al Moustafa et al. observed co-over expression of E6/E7 and HER-2 in 40% of HPV16 positive BC (99). Ohba et al. showed association of the APOBEC3B pathway with the ER-positive breast tumors in presence of HPV (56). The association of E6 with these pathways in breast carcinogenesis has been validated in murine model systems (100).
Future management of HPV associated BC
In this review, it is evident that HPV is associated with a sub set of BC irrespective of different molecular subtypes. As HPV infects the breast through nipple and micro-lesions on the breast skin due to genetial-breast sex activity, hygienic sexual practice could prevent HPV infection to the breast. In conventional cervical cancer screening, cervical swab is used for HPV test followed by Pap test leading to early diagnosis of cervical cancer (101). Likewise, it is pertinent to detect HPV in breast ductal lavage, breast nipple discharge and breast milk which will be useful for determination of risk of BC as well as early diagnosis of BC. Apart from these, detection of HPV in breast tissue will be powerful biomarker for specific treatment protocol of the HPV infected BC. Moreover, the presence of HPV in blood plasma of BC patients can be the indicator of dissemination of tumour cells from the primary site which can serve as a useful prognostic tool of the disease. The prevalence of HPV in BC indicates that prophylactic vaccination against HPV is needed to restrict the disease in women (102).
Conclusions
In this review, we suggest that HPV is an important etiological factor in the development of a sub-set of BC and also HPV associated BC has some distinct molecular profile than other HPV associated cancers like cervical cancer (CACX), head and neck squamous cell carcinoma (HNSCC). Thus an in-depth understanding and analysis of the molecular profile of BC in the light of HPV is essentially needed for the proper management of the disease.
Acknowledgments
The authors thank the Director, Chittaranjan National Cancer Institute, Kolkata, India for kind interest in the work. We would like to thank Mr. Aniban Roychowdhury for his language editing help and valuable suggestions.
Funding: The financial support for this work was provided by UGC-NET Fellowship grant Sr. No. 2121430433, Ref. No.: 21/12/2014(ii) EU-V dated 08.06.2015 to Mr. BC.
Footnote
Conflict of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-19-2756). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Malvia S, Bagadi SA, Dubey US, et al. Epidemiology of breast cancer in Indian women. Asia Pac J Clin Oncol 2017;13:289-95. [Crossref] [PubMed]
- Hankinson SE, Colditz GA, Willett WC. Towards an integrated model for breast cancer etiology: the lifelong interplay of genes, lifestyle, and hormones. Breast Cancer Res 2004;6:213-8. [Crossref] [PubMed]
- Alibek K, Kakpenova A, Mussabekova A, et al. Role of viruses in the development of breast cancer. Infect Agent Cancer 2013;8:32. [Crossref] [PubMed]
- de Lima EG, do Amaral CM, Peixe FC, et al. Putative Mechanisms of Viral Transmission and Molecular Dysregulation of Mammary Epithelial Cells by Human Papillomavirus: Implications for Breast Cancer. Curr Mol Med 2016. [Epub ahead of print]. [PubMed]
- Wang T, Chang P, Wang L, et al. The role of human papillomavirus infection in breast cancer. Med Oncol 2012;29:48-55. [Crossref] [PubMed]
- Li J, Ding J, Zhai K. Detection of Human Papillomavirus DNA in Patients with Breast Tumor in China. PLoS One 2015;10:e0136050. [Crossref] [PubMed]
- de Villiers EM, Sandstrom RE, zur Hausen H, et al. Presence of papillomavirus sequences in condylomatous lesions of the mamillae and in invasive carcinoma of the breast. Breast Cancer Res 2005;7:R1-11. [Crossref] [PubMed]
- Polyak K. Breast cancer: origins and evolution. J Clin Invest 2007;117:3155-63. [Crossref] [PubMed]
- Ma H, Wang Y, Sullivan-Halley J, et al. Use of four biomarkers to evaluate the risk of breast cancer subtypes in the women's contraceptive and reproductive experiences study. Cancer Res 2010;70:575-87. [Crossref] [PubMed]
- Yu Y, Morimoto T, Sasa M, et al. HPV33 DNA in premalignant and malignant breast lesions in Chinese and Japanese populations. Anticancer Res 1999;19:5057-61. [PubMed]
- Yu Y, Morimoto T, Sasa M, et al. Human papillomavirus type 33 DNA in breast cancer in Chinese. Breast Cancer 2000;7:33-6. [Crossref] [PubMed]
- Damin AP, Karam R, Zettler CG, et al. Evidence for an association of human papillomavirus and breast carcinomas. Breast Cancer Res Treat 2004;84:131-7. [Crossref] [PubMed]
- Gumus M, Yumuk PF, Salepci T, et al. HPV DNA frequency and subset analysis in human breast cancer patients' normal and tumoral tissue samples. J Exp Clin Cancer Res 2006;25:515-21. [PubMed]
- Kroupis C, Markou A, Vourlidis N, et al. Presence of high-risk human papillomavirus sequences in breast cancer tissues and association with histopathological characteristics. Clin Biochem 2006;39:727-31. [Crossref] [PubMed]
- Choi YL, Cho EY, Kim JH, et al. Detection of human papillomavirus DNA by DNA chip in breast carcinomas of Korean women. Tumour Biol 2007;28:327-32. [Crossref] [PubMed]
- Tsai JH, Tsai CH, Cheng MH, et al. Association of viral factors with non-familial breast cancer in Taiwan by comparison with non-cancerous, fibroadenoma, and thyroid tumor tissues. J Med Virol 2005;75:276-81. [Crossref] [PubMed]
- Khan NA, Castillo A, Koriyama C, et al. Human papillomavirus detected in female breast carcinomas in Japan. Br J Cancer 2008;99:408-14. [Crossref] [PubMed]
- de León DC, Montiel DP, Nemcova J, et al. Human papillomavirus (HPV) in breast tumors: prevalence in a group of Mexican patients. BMC Cancer 2009;9:26. [Crossref] [PubMed]
- Heng B, Glenn WK, Ye Y, et al. Human papilloma virus is associated with breast cancer. Br J Cancer 2009;101:1345-50. [Crossref] [PubMed]
- He Q, Zhang SQ, Chu YL, et al. The correlations between HPV16 infection and expressions of c-erbB-2 and bcl-2 in breast carcinoma. Mol Biol Rep 2009;36:807-12. [Crossref] [PubMed]
- Mendizabal-Ruiz AP, Morales JA, Ramirez-Jirano LJ, et al. Low frequency of human papillomavirus DNA in breast cancer tissue. Breast Cancer Res Treat 2009;114:189-94. [Crossref] [PubMed]
- Herrera-Goepfert R, Khan NA, Koriyama C, et al. High-risk human papillomavirus in mammary gland carcinomas and non-neoplastic tissues of Mexican women: no evidence supporting a cause and effect relationship. Breast 2011;20:184-9. [Crossref] [PubMed]
- Mou X, Chen L, Liu F, et al. Low prevalence of human papillomavirus (HPV) in Chinese patients with breast cancer. J Int Med Res 2011;39:1636-44. [Crossref] [PubMed]
- Frega A, Lorenzon L, Bononi M, et al. Evaluation of E6 and E7 mRNA expression in HPV DNA positive breast cancer. Eur J Gynaecol Oncol 2012;33:164-7. [PubMed]
- Glenn WK, Heng B, Delprado W, et al. Epstein-Barr virus, human papillomavirus and mouse mammary tumour virus as multiple viruses in breast cancer. PLoS One 2012;7:e48788. [Crossref] [PubMed]
- Sigaroodi A, Nadji SA, Naghshvar F, et al. Human papillomavirus is associated with breast cancer in the north part of Iran. ScientificWorldJournal 2012;2012:837191. [Crossref] [PubMed]
- Liang W, Wang J, Wang C, et al. Detection of high-risk human papillomaviruses in fresh breast cancer samples using the hybrid capture 2 assay. J Med Virol 2013;85:2087-92. [Crossref] [PubMed]
- Wang T, Zeng X, Li W, et al. Detection and analysis of human papillomavirus (HPV) DNA in breast cancer patients by an effective method of HPV capture. PLoS One 2014;9:e90343. [Crossref] [PubMed]
- Ali SH, Al-Alwan NA, Al-Alwany SH. Detection and genotyping of human papillomavirus in breast cancer tissues from Iraqi patients. East Mediterr Health J 2014;20:372-7. [Crossref] [PubMed]
- Ahangar-Oskouee M, Shahmahmoodi S, Jalilvand S, et al. No detection of 'high-risk' human papillomaviruses in a group of Iranian women with breast cancer. Asian Pac J Cancer Prev 2014;15:4061-5. [Crossref] [PubMed]
- Manzouri L, Salehi R, Shariatpanahi S, et al. Prevalence of human papilloma virus among women with breast cancer since 2005-2009 in Isfahan. Adv Biomed Res 2014;3:75. [Crossref] [PubMed]
- Peng J, Wang T, Zhu H, et al. Multiplex PCR/mass spectrometry screening of biological carcinogenic agents in human mammary tumors. J Clin Virol 2014;61:255-9. [Crossref] [PubMed]
- Fu L, Wang D, Shah W, et al. Association of human papillomavirus type 58 with breast cancer in Shaanxi province of China. J Med Virol 2015;87:1034-40. [Crossref] [PubMed]
- Lawson JS, Glenn WK, Salyakina D, et al. Human Papilloma Viruses and Breast Cancer. Front Oncol 2015;5:277. [Crossref] [PubMed]
- Ngan C, Lawson JS, Clay R, et al. Early Human Papilloma Virus (HPV) Oncogenic Influences in Breast Cancer. Breast Cancer (Auckl) 2015;9:93-7. [Crossref] [PubMed]
- Salman NA, Davies G, Majidy F, et al. Association of High Risk Human Papillomavirus and Breast cancer: A UK based Study. Sci Rep 2017;7:43591. [Crossref] [PubMed]
- Islam S, Dasgupta H, Roychowdhury A, et al. Study of association and molecular analysis of human papillomavirus in breast cancer of Indian patients: Clinical and prognostic implication. PLoS One 2017;12:e0172760. [Crossref] [PubMed]
- Delgado-García S, Martinez-Escoriza JC, Alba A, et al. Presence of human papillomavirus DNA in breast cancer: a Spanish case-control study. BMC Cancer 2017;17:320. [Crossref] [PubMed]
- Khodabandehlou N, Mostafaei S, Etemadi A, et al. Human papilloma virus and breast cancer: the role of inflammation and viral expressed proteins. BMC Cancer 2019;19:61. [Crossref] [PubMed]
- Wrede D, Luqmani YA, Coombes RC, et al. Absence of HPV 16 and 18 DNA in breast cancer. Br J Cancer 1992;65:891-4. [Crossref] [PubMed]
- Bratthauer GL, Tavassoli FA, O'Leary TJ. Etiology of breast carcinoma: no apparent role for papillomavirus types 6/11/16/18. Pathol Res Pract 1992;188:384-6. [Crossref] [PubMed]
- Gopalkrishna V, Singh UR, Sodhani P, et al. Absence of human papillomavirus DNA in breast cancer as revealed by polymerase chain reaction. Breast Cancer Res Treat 1996;39:197-202. [Crossref] [PubMed]
- Lindel K, Forster A, Altermatt HJ, et al. Breast cancer and human papillomavirus (HPV) infection: no evidence of a viral etiology in a group of Swiss women. Breast 2007;16:172-7. [Crossref] [PubMed]
- de Cremoux P, Thioux M, Lebigot I, et al. No evidence of human papillomavirus DNA sequences in invasive breast carcinoma. Breast Cancer Res Treat 2008;109:55-8. [Crossref] [PubMed]
- Chang P, Wang T, Yao Q, et al. Absence of human papillomavirus in patients with breast cancer in north-west China. Med Oncol 2012;29:521-5. [Crossref] [PubMed]
- Lüder Ripoli F, Mohr A, Conradine Hammer S, et al. A Comparison of Fresh Frozen vs. Formalin-Fixed, Paraffin-Embedded Specimens of Canine Mammary Tumors via Branched-DNA Assay. Int J Mol Sci 2016;17:724. [Crossref] [PubMed]
- Cavalcante JR, Pinheiro LGP, Almeida PRC, et al. Association of breast cancer with human papillomavirus (HPV) infection in Northeast Brazil: molecular evidence. Clinics (Sao Paulo) 2018;73:e465. [Crossref] [PubMed]
- Ngamkham J, Karalak A, Chaiwerawattana A, et al. Prevalence of Human Papillomavirus Infection in Breast Cancer Cells from Thai Women. Asian Pac J Cancer Prev 2017;18:1839-45. [PubMed]
- Corbex M, Bouzbid S, Traverse-Glehen A, et al. Prevalence of papillomaviruses, polyomaviruses, and herpesviruses in triple-negative and inflammatory breast tumors from algeria compared with other types of breast cancer tumors. PLoS One 2014;9:e114559. [Crossref] [PubMed]
- Piana AF, Sotgiu G, Muroni MR, et al. HPV infection and triple-negative breast cancers: an Italian case-control study. Virol J 2014;11:190. [Crossref] [PubMed]
- Vernet-Tomas M, Mena M, Alemany L, et al. Human papillomavirus and breast cancer: no evidence of association in a Spanish set of cases. Anticancer Res 2015;35:851-6. [PubMed]
- Fernandes A, Bianchi G, Feltri AP, et al. Presence of human papillomavirus in breast cancer and its association with prognostic factors. Ecancermedicalscience 2015;9:548. [Crossref] [PubMed]
- Habyarimana T, Attaleb M, Mazarati JB, et al. Detection of human papillomavirus DNA in tumors from Rwandese breast cancer patients. Breast Cancer 2018;25:127-33. [Crossref] [PubMed]
- Woods Ignatoski KM, Dziubinski ML, Ammerman C, et al. Cooperative interactions of HER-2 and HPV-16 oncoproteins in the malignant transformation of human mammary epithelial cells. Neoplasia 2005;7:788-98. [Crossref] [PubMed]
- Ohba K, Ichiyama K, Yajima M, et al. In vivo and in vitro studies suggest a possible involvement of HPV infection in the early stage of breast carcinogenesis via APOBEC3B induction. PLoS One 2014;9:e97787. [Crossref] [PubMed]
- Bae JM, Kim EH. Human papillomavirus infection and risk of breast cancer: a meta-analysis of case-control studies. Infect Agent Cancer 2016;11:14. [Crossref] [PubMed]
- Carolis S, Pellegrini A, Santini D, et al. Liquid biopsy in the diagnosis of HPV DNA in breast lesions. Future Microbiol 2018;13:187-94. [Crossref] [PubMed]
- Balci FL, Uras C, Feldman SM. Is human papillomavirus associated with breast cancer or papilloma presenting with pathologic nipple discharge? Cancer Treat Res Commun 2019;19:100122. [Crossref] [PubMed]
- Louvanto K, Sarkola M, Rintala M, et al. Breast Milk Is a Potential Vehicle for Human Papillomavirus Transmission to Oral Mucosa of the Spouse. Pediatr Infect Dis J 2017;36:627-30. [Crossref] [PubMed]
- Tuominen H, Rautava S, Collado MC, et al. HPV infection and bacterial microbiota in breast milk and infant oral mucosa. PLoS One 2018;13:e0207016. [Crossref] [PubMed]
- Diaz S, Boulle N, Moles JP, et al. Human papillomavirus (HPV) shedding in breast milk from African women living with HIV. J Clin Virol 2018;106:41-3. [Crossref] [PubMed]
- Stevens-Simon C, Nelligan D, Breese P, et al. The prevalence of genital human papillomavirus infections in abused and nonabused preadolescent girls. Pediatrics 2000;106:645-9. [Crossref] [PubMed]
- Beutner KR, Wiley DJ, Douglas JM, et al. Genital warts and their treatment. Clin Infect Dis 1999;28 Suppl 1:S37-56. [Crossref] [PubMed]
- Giroglou T, Florin L, Schafer F, et al. Human papillomavirus infection requires cell surface heparan sulfate. J Virol 2001;75:1565-70. [Crossref] [PubMed]
- Joyce JG, Tung JS, Przysiecki CT, et al. The L1 major capsid protein of human papillomavirus type 11 recombinant virus-like particles interacts with heparin and cell-surface glycosaminoglycans on human keratinocytes. J Biol Chem 1999;274:5810-22. [Crossref] [PubMed]
- Culp TD, Christensen ND. Kinetics of in vitro adsorption and entry of papillomavirus virions. Virology 2004;319:152-61. [Crossref] [PubMed]
- Day PM, Lowy DR, Schiller JT. Papillomaviruses infect cells via a clathrin-dependent pathway. Virology 2003;307:1-11. [Crossref] [PubMed]
- Selinka HC, Giroglou T, Sapp M. Analysis of the infectious entry pathway of human papillomavirus type 33 pseudovirions. Virology 2002;299:279-87. [Crossref] [PubMed]
- Li M, Beard P, Estes PA, et al. Intercapsomeric disulfide bonds in papillomavirus assembly and disassembly. J Virol 1998;72:2160-7. [Crossref] [PubMed]
- Lawson JS, Glenn WK, Salyakina D, et al. Human Papilloma Virus Identification in Breast Cancer Patients with Previous Cervical Neoplasia. Front Oncol 2016;5:298. [Crossref] [PubMed]
- Widschwendter A, Brunhuber T, Wiedemair A, et al. Detection of human papillomavirus DNA in breast cancer of patients with cervical cancer history. J Clin Virol 2004;31:292-7. [Crossref] [PubMed]
- Hennig EM, Suo Z, Thoresen S, et al. Human papillomavirus 16 in breast cancer of women treated for high grade cervical intraepithelial neoplasia (CIN III). Breast Cancer Res Treat 1999;53:121-35. [Crossref] [PubMed]
- Bodaghi S, Wood LV, Roby G, et al. Could human papillomaviruses be spread through blood? J Clin Microbiol 2005;43:5428-34. [Crossref] [PubMed]
- Islam S, Dasgupta H, Basu M, et al. Skin mediates Human Papilloma Virus (HPV) infection in breast: A report of four cases. Available online: https://www.researchgate.net/publication/324008020_Skin_mediated_human_papillomavirus_infection_in_breast_A_report_of_four_cases
- Breast cancer may be sexually transmitted. 2006. Available online: www.abc.net.au/science/news/stories/2006/1808903.htm. Accessed 12 December.
- Wallin KL, Wiklund F, Angstrom T, et al. Type-specific persistence of human papillomavirus DNA before the development of invasive cervical cancer. N Engl J Med 1999;341:1633-8. [Crossref] [PubMed]
- Zielinski GD, Snijders PJ, Rozendaal L, et al. HPV presence precedes abnormal cytology in women developing cervical cancer and signals false negative smears. Br J Cancer 2001;85:398-404. [Crossref] [PubMed]
- zur Hausen H. Papillomavirus infections--a major cause of human cancers. Biochim Biophys Acta 1996;1288:F55-78. [PubMed]
- zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer 2002;2:342-50. [Crossref] [PubMed]
- Aguayo F, Khan N, Koriyama C, et al. Human papillomavirus and Epstein-Barr virus infections in breast cancer from chile. Infect Agent Cancer 2011;6:7. [Crossref] [PubMed]
- Herrera-Goepfert R, Vela-Chavez T, Carrillo-Garcia A, et al. High-risk human papillomavirus (HPV) DNA sequences in metaplastic breast carcinomas of Mexican women. BMC Cancer 2013;13:445. [Crossref] [PubMed]
- Lawson JS, Glenn WK, Whitaker NJ. Human Papilloma Viruses and Breast Cancer - Assessment of Causality. Front Oncol 2016;6:207. [Crossref] [PubMed]
- Pereira Suarez AL, Lorenzetti MA, Gonzalez Lucano R, et al. Presence of human papilloma virus in a series of breast carcinoma from Argentina. PLoS One 2013;8:e61613. [Crossref] [PubMed]
- McBride AA, Warburton A. The role of integration in oncogenic progression of HPV-associated cancers. PLoS Pathog 2017;13:e1006211. [Crossref] [PubMed]
- Burk RD, Harari A, Chen Z. Human papillomavirus genome variants. Virology 2013;445:232-43. [Crossref] [PubMed]
- Mirabello L, Yeager M, Cullen M, et al. HPV16 Sublineage Associations With Histology-Specific Cancer Risk Using HPV Whole-Genome Sequences in 3200 Women. J Natl Cancer Inst 2016;108:djw100. [Crossref] [PubMed]
- DeFilippis VR, Ayala FJ, Villarreal LP. Evidence of diversifying selection in human papillomavirus type 16 E6 but not E7 oncogenes. J Mol Evol 2002;55:491-9. [Crossref] [PubMed]
- Doorbar J, Egawa N, Griffin H, et al. Human papillomavirus molecular biology and disease association. Rev Med Virol 2015;25 Suppl 1:2-23. [Crossref] [PubMed]
- Johansson C, Schwartz S. Regulation of human papillomavirus gene expression by splicing and polyadenylation. Nat Rev Microbiol 2013;11:239-51. [Crossref] [PubMed]
- Balasubramaniam SD, Balakrishnan V, Oon CE, et al. Key Molecular Events in Cervical Cancer Development. Medicina (Kaunas) 2019;55:384. [Crossref] [PubMed]
- Wazer DE, Liu XL, Chu Q, et al. Immortalization of distinct human mammary epithelial cell types by human papilloma virus 16 E6 or E7. Proc Natl Acad Sci U S A 1995;92:3687-91. [Crossref] [PubMed]
- Liu Y, Chen JJ, Gao Q, et al. Multiple functions of human papillomavirus type 16 E6 contribute to the immortalization of mammary epithelial cells. J Virol 1999;73:7297-307. [Crossref] [PubMed]
- Wang YX, Zhang ZY, Wang JQ, et al. HPV16 E7 increases COX-2 expression and promotes the proliferation of breast cancer. Oncol Lett 2018;16:317-25. [PubMed]
- Zhang Y, Fan S, Meng Q, et al. BRCA1 interaction with human papillomavirus oncoproteins. J Biol Chem 2005;280:33165-77. [Crossref] [PubMed]
- Rosen EM, Fan S, Isaacs C. BRCA1 in hormonal carcinogenesis: basic and clinical research. Endocr Relat Cancer 2005;12:533-48. [Crossref] [PubMed]
- Hilakivi-Clarke L. Estrogens, BRCA1, and breast cancer. Cancer Res 2000;60:4993-5001. [PubMed]
- Yasmeen A, Bismar TA, Kandouz M, et al. E6/E7 of HPV type 16 promotes cell invasion and metastasis of human breast cancer cells. Cell Cycle 2007;6:2038-42. [Crossref] [PubMed]
- Al Moustafa AE, Kassab A, Darnel A, et al. High-risk HPV/ErbB-2 interaction on E-cadherin/catenin regulation in human carcinogenesis. Curr Pharm Des 2008;14:2159-72. [Crossref] [PubMed]
- Shai A, Pitot HC, Lambert PF. p53 Loss synergizes with estrogen and papillomaviral oncogenes to induce cervical and breast cancers. Cancer Res 2008;68:2622-31. [Crossref] [PubMed]
- Koliopoulos G, Nyaga VN, Santesso N, et al. Cytology versus HPV testing for cervical cancer screening in the general population. Cochrane Database Syst Rev 2017;8:CD008587. [PubMed]
- Purdie J. Can Human Papillomavirus (HPV) Cause Breast Cancer? healthline. 2018. Available online: https://www.healthline.com/health/breast-cancer/breast-cancer-and-hpv. Accessed December 14 2018.


