
The application of weighted gene co-expression network analysis in identifying key modules and hub genes associated with disease status in Alzheimer’s disease
Introduction
Alzheimer’s disease (AD), as the most common neurodegenerative condition, affects more than 15 million people worldwide (1,2) and is characterized by progressive memory loss and cognitive deterioration (1,3). Deposits of extracellular amyloid plaques made of amyloid-β and neuronal intracellular neurofibrillary tangles (NFTs) composed of the protein tau are the primary pathological manifestations of AD (4).
AD can be divided into six stages, called Braak stages, based on the site of the tangle-bearing neurons and the rigorousness of modifications. The stages are defined as follows: I–II (transentorhinal stages), clinically silent cases; III–IV (limbic stage), incipient AD; and V–VI (neocortical stages), fully developed AD (3,5). The mini-mental state examination (MMSE) is a cognitive test used to screen and monitor dementia progression, such as that found in AD. It has been reported that a score below 24 of a possible 30 is indicative of dementia (6), and non-demented elderly patients usually score outside this range at 24 or above. The NFT scoring system is another related measure that is usually in accordance with Braak stages, with NFT III stage being indicative of AD (7).
Many researchers have found comprehensive evidence of the molecular pathogenesis of AD, and this information has been converted into promising treatment approaches, including immunotherapy using amyloid‐β, which is being tested clinically (8). To understand the biology of AD and to provide measures of relevant pathophysiology in living individuals, biomarkers are becoming increasingly important in both AD patients and pre-symptomatic individuals. For AD patients, the biomarkers in the cerebrospinal fluid (CSF) are divided into basic and core biomarkers (9). Basic biomarkers comprise assays for blood-brain barrier (BBB) status and inflammatory methods, which have been developed to recognize disorders that may mimic or coincide with AD, for example, neuroborreliosis. Meanwhile, core biomarkers are used to recognize the chief pathogenic progressions of AD patients, and mainly reflect NFT and amyloid pathology, or axonal degeneration, such as Aβ1–42 (10,11), total tau levels (12,13), and phosphorylated tau levels (phosphorylated at thr181 or thr231) (14,15). Apart from aβ and tau, many other novel candidate biomarkers have been recognized for CSF biomarkers (16). These included βsite APP cleaving enzyme 1 (BACE1), which is the chief enzyme accountable for β-secretase activity; amyloid-β oligomers, the accumulation of which can form insoluble fibrillar Aβ in plaques; truncated amyloid-β isoforms, the most available aβ isoform in the CSF; and neuronal and synaptic markers, such as visinin-like protein 1, synaptotagmin, and growth-associated protein (9). It has been a challenge for independent studies to confirm the role of biomarkers in AD in peripheral blood; however, numerous candidate blood biomarkers have been anticipated.
Traditional studies of the mechanisms and biomarkers of AD predominantly emphasize a single gene or a specific pathway. However, the growth and advancement of AD is a complex process that is controlled by the accumulative impact of a sequence of genes and their interrelations.
As per the theory that genes with an analogous expression pattern might have same functions or take part in common cascades (17), the weighted gene co-expression network analysis (WGCNA) has emerged as an active approach for understanding the associations among diverse gene sets or among gene sets and clinical features (18,19). WGCNA has been extensively utilized in finding the hub genes related to the clinical features of various diseases (20-22).
In this study, WGCNA was implemented to conjointly examine clinical evidence and microarray records of AD patients’ samples for the purpose of recognizing vital genes related to medical features. These vital genes may have essential clinical implications and be informative as investigative and predictive biomarkers or treatment targets. Finally, one of the hub genes, glutamate ionotropic receptor kainate type subunit 1 (GRIK1), was validated by an animal AD model, with the results showing that the mRNA and protein level of GRIK1 in the AD hippocampus was increased compared with control (NC) hippocampus. Phalloidin staining showed that dendritic length of GRIK1 pCDNA3.1 group was shorter than the NC group.
Methods
Data collection and preprocessing
Two expression profile data sets of AD were acquired from the Gene Expression Omnibus (GEO) database. Datasets GSE1297 and GSE28146, which focus on critical early stages, were chosen for further study (23,24). The GSE1297 dataset was utilized for AD-correlated modules and gene selection. Another dataset was used for independent verification.
Probes were mapped to gene symbols. Probes with more than one gene and empty probes were removed as per the annotation platform of each expression profile. If there were numerous probes, which mapped to the similar gene symbol, their mean value was regarded as the gene manifestation value. Consequently, 12,502 unique genes representing the expression profiles of GSE1297, and 20846 unique genes representing GSE28146, were used for analysis.
In addition, as a common challenge in examining genome-wide expression data is managing batch effects, it is necessary to continuously monitor for batch effects whenever entirely analogous processing of samples is impossible. These data were normalized by means of the normalizeBetweenArrays() function in the limma library in R. After screening out the genes with the highest median absolute deviation (MAD) of 75%, 9,974 genes remained from GSE1297.
Co-expression module detection
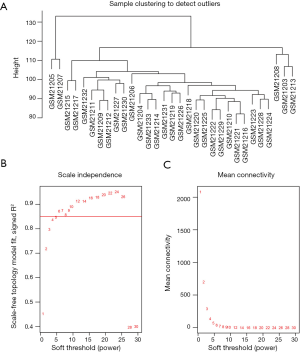
The hclust() function in the stats library in the R was used to perform cluster analysis of the samples with the suitable threshold value to both identify and eliminate the outliers. The gradient method was utilized to examine the independence and the average degree of connectivity of the numerous modules with diverse power values (the power values oscillated from 1 to 30). Once the suitable power value had been recognized when the degree of independence was 0.85, the module creation continued with the WGCNA process. Module identification was achieved by means of the dynamic tree cut method. The least number of genes was fixed at 30 to confirm greater dependability. Successively, the information relating to the analogous genes in each module was obtained.
Module and clinical trait association analysis
The WGCNA algorithm uses module eigengenes (MEs) to evaluate the possible relationship of gene modules with clinical traits. MEs were defined as the primary key constituents computed by means of principal component analysis that recapitulates the manifestation of genes of a specific module into a single characteristic expression profile. The manifestation configurations of modules related to the types of samples were quantified by gene significance (GS) and module significance (MS). The GS measure was defined as the value of the Pearson correlation among the i-th gene profile xi and the sample trait T:
| [1] |
MS was defined as the average GS for all the genes in the module.
Gene ontology (GO) and pathway enrichment analyses
The GO enrichment analyses of concerned gene sets were accomplished by means of DAVID (25).
Association analysis and hub genes
The ME-based connectivity measure (kme) is the distance from the expression profile of a gene to that of the ME. The ME provides upsurge to a measure of module centrality (kme) as mentioned below:
| [2] |
x(i) is the profile of gene i and ME(q) is the ME of module q. Therefore, kme computes how adjacent a gene is to a module; i.e., it assesses the module membership of a gene. The hub genes are the ones with extensive network connectivity in a specific group. Moreover, the hub genes of modules are also largely related to the analogous clinical traits of the modules. Hence, genes with a high kme in the module were casually denoted as intramodular hub genes in this study.
Efficacy evaluation
Receiver operating characteristic (ROC) curve analysis was used to investigate the value of hub genes in diagnostic efficiency to differentiate among severe and non-severe AD patients. Detailed information of patients was obtained from a description of the GSE28146 dataset. The result was confirmed by the GSE28146 validation set. To do this, logistic regression was employed to classify the samples as follows:
| [3] |
Where p is the probability of severe stage, β0…βk are the parameters to be estimated, and k is number of genes in a given module.
ROC curve was schemed, and the area under the curve (AUC) was quantified with “ROCR” package (Sing T, et al., 2005; Bioinformatics). When an AUC value was higher than 0.7, the hub gene was regarded as having outstanding specificity and sensitivity. The outcome was established by the GSE28146 validation set.
Animals
The AD mice and wild type (WT) mice we used were bought from Charles River (China). Briefly, the human mutant amyloid precursor protein (APP) (HuAPPsw) (Tg2576) transgenic mouse AD model was used. Animal experiments were conducted in accordance with the guideline for the Care and Use of Laboratory Animals published by the US National Institutes of Health.
Total RNA extraction and quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) analysis
Total RNA was isolated from AD mice and WT littermates (NC) using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. The RNA of the cortex and hippocampus was isolated separately from the two groups. qRT-PCR was employed to determine the mRNA expression of GRLK1. qRT-PCR was performed using SYBRGreen qPCR Master Mix (TAKARA). The primer sequences are as follows: GRIK1 forward: 5-CCGGGAATTCCATGTTTTGTGATAGTTTTGCA-3, GRIK1 reverse: 5-GAGTTCCTCGAGTCAGCTATGGTTTTGATCTT-3; β-actin forward: 5-CCCATCTATGAGGGTTACGC-3, β-actin reverse: 5-TTTAATGTCACGCACGATTTC-3.
Western blot analysis
Tissue was extracted using cell lysis buffer followed by immunoblotting with anti-GRIK1 (Abcam) and anti-β-actin (Santa Cruz Biotechnology). Cells were lysed in RIPA buffer with protease inhibitors (Roche Applied Science) on ice for 20 min and then centrifuged at 13,500 rpm for 20 min at 4 °C. The supernatant was placed into a new tube, and the protein concentration was measured using the bicinchoninic acid protein assay kit (Applygen). Next, 30 µg of cell lysates were resolved with 12.5% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. Then, the membranes were blocked with 5% skim milk for 1 h at room temperature (RT). After blocking, the membranes were incubated with primary antibody for 1 h at RT. Then, the membranes were incubated with secondary antibodies at RT for 1 h. The protein bands were visualized using enhanced chemiluminescence chromogenic substrate with horseradish peroxidase (Beijing, China).
Primary neuron culture
Primary neurons were isolated from the hippocampus of neonatal mice (26). Briefly, the extracted hippocampus was digested in 0.25% trypsin for 20 min. Cells were collected by centrifugation at 200 ×g for 5 min. Isolated cells were cultured and maintained in NeurobasalTM medium (GIBCO) supplemented with B27 and L-glutamine (GIBCO).
Cell transfection
The sequence of GRIK1 used human liver cDNA (Invitrogen) and was cloned into the pCDNA3.1 vector (GRIK1 pCDNA3.1). Primary neurons were seeded into a six-well plate at a concentration of 105, and the plasmids or its control plasmids were transfected using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s protocols.
Phalloidin staining
Primary neurons were fixed in 4% PFA for 20 min, permeabilized using 0.5% Triton X-100 (Biosharp, China) for 30 min, and treated with Alexa Fluor® 488 conjugated phalloidin (Cell Signaling, USA) for 30 min and the nuclear counterstain DAPI for 5 min. Coverslips were washed and mounted on glass slides. Stained neurons were photographed using a Nikon C2si confocal microscope (Nikon) equipped with a 60× oil immersion objective.
Results
Identification of gene co-expression modules
To determine if all 31 samples in GSE1297 were appropriate for network analysis, the sample dendrogram and analogous clinical traits were investigated. We confirmed that all samples were incorporated into the groups and conceded the cutoff thresholds (Figure 1A). Soft-thresholding power is a crucial factor for creating a WGCNA. We accomplished the analysis of network topology for thresholding powers from 1 to 20. Figure 1A,B,C demonstrate that when the power value was equivalent to 6 (scale-free R2 =0.872), our records projected a gene co-expression network that displayed scale-free topology with integral modular features.
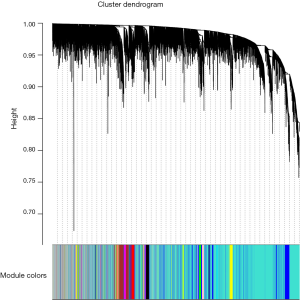
Finally, we created average linkage hierarchical clustering with the topological overlap matrix (TOM) and created the gene co-expression network by means of WGCNA depending on the graded grouping of the quantified variations. Subsequently, 16 modules were acquired (Figure 2). We utilized MEs as illustrative profiles and computed module correspondence by eigengene correlation. Interaction relations of the 16 modules were examined, and the network heatmap was designed (Figure 3A). The outcomes indicated that each module was autonomously authenticated with another which revealed a high level of individuality among the modules and the comparative independence of gene manifestation in each module.
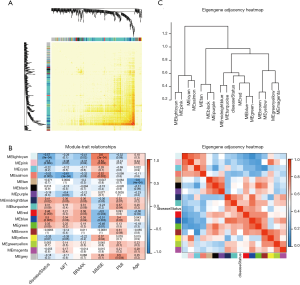
Correlation of modules and clinical traits
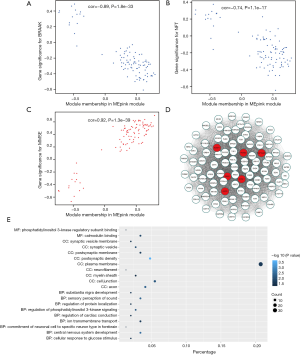
To investigate if any module was connected with disease status, we investigated the relevance among each module plus the AD clinical traits, including MMSE, BRAAK, NFT, postmortem interval (PMI), and age. We found that some modules were highly correlated with the disease status of AD (modules of lightcyan, pink, salmon, and blue). For example, the ME value of the lightcyan module also exhibited a positive association with MMSE (r=0.6, P=3×10−4) and a negative association with BRAAK (r=−0.41, P=0.02) (Figure 3B). The eigengene dendrogram and the heatmap specified that the 16 modules were mostly separated into two groups (Figure 3C). Moreover, the ME of the pink module indicated a significant connection with all disease status (NFT, BRAAK, and MMSE) compare to the other modules (Figure 3B), indicating that the pink module might have an integral role in the occurrence and development of AD. We recognized the pink module as the module most applicable to the disease level of AD. Finally, a correlation between module membership and GS was plotted for the module pink (Figure 4A,B,C). We visualized the pink module as a network in Cytoscape. Figure 4D shows the genes in the pink module, and the red nodes signify hub genes. The hub genes in the pink module comprised acetoacetyl-CoA synthetase (AACS), GRIK1, homeobox B2 (HOXB2), potassium voltage-gated channel modifier subfamily F member 1 (KCNF1), MYB proto-oncogene like 1 (MYBL1), and RAP1 GTPase activating protein 2 (RAP1GAP2).
Functional enrichment analysis of genes in modules of interest
Since genes with analogous expression patterns might take part in parallel biological procedures or networks, we achieved enrichment analysis to search the GO terms in which the pink module was incorporated. As per the obtained outcomes, the pink module was mostly enriched in central nervous system development, regulation of phosphatidylinositol 3-kinase (PI3K) signaling, and ion transmembrane transport (Figure 4E & Table 1). To further investigate the role of modules in AD, we also performed GO enrichment analysis to other modules, which were correlated with two of three clinical traits (modules of lightcyan, salmon, and blue). Interestingly, the genes in the blue, salmon and pink modules were significantly enriched in the GO terms of the plasma membrane (Figure S1).
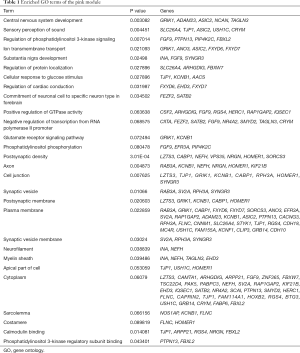
Full table
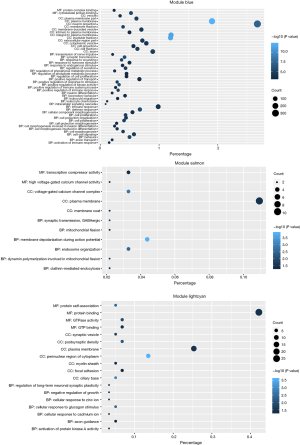
Efficacy evaluation of hub genes
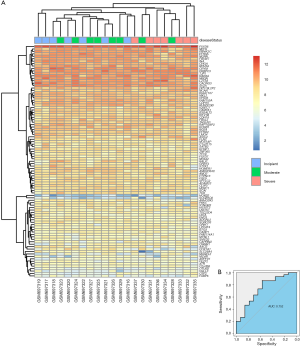
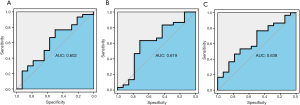
In the GSE28146 validation set, we found that genes in the pink module are capable of discriminating severe and non-severe AD samples (Figure 5A). Furthermore, ROC curve analysis was executed to assess the diagnostic effectiveness of hub genes to differentiate among severe and non-severe AD samples. We quantified the AUC value and designed ROC curves for the six hub genes. The AUC value of the pink module was greater than 0.7 (Figure 5B). The ROC curves for hub genes of the other less related modules (modules of lightcyan, salmon, and blue) are presented in Figure S2.
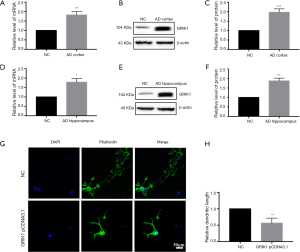
GRIK1 was upregulated in the AD cortex and AD hippocampus and attenuated dendritic length in primary neurons
To identify the levels of GRIK1 in the AD cortex and AD hippocampus, qRT-PCR and western blot were used. The qRT-PCR data showed that the mRNA level of GRIK1 was higher in the AD cortex compared with the NC cortex (Figure 6A). Western blotting data showed that the expression of GRIK1 protein was higher in the AD cortex compared to the NC cortex (Figure 6B,C). A comparison of the expression of GRIK1 between the AD hippocampus and NC hippocampus was also examined via qRT-PCR and Western blot. The mRNA level of GRIK1 in the AD hippocampus was increased compared with the NC hippocampus (Figure 6D). The protein level of GRIK1 in the AD hippocampus showed the same pattern (Figure 6E,F). Phalloidin staining showed that the dendritic length of GRIK1 pCDNA3.1 group was shorter than that in the NC group (Figure 6G,H), These data indicated that GRIK1 was upregulated both in the AD cortex and AD hippocampus and attenuated the dendritic length of primary neurons.
Discussion
This study combined the mining of microarray data with the clinical evidence of AD patients to perform a WGCNA with the aim of recognizing those biomarkers related to clinical features that could distinguish the different stages of AD patients.
There are many different stage standards for differentiating AD types. Based on the foundation of the dispersal of NFTs, AD can be divided into three subtypes (Braak stage ≥ IV): hippocampal-sparing AD, typical AD, and limbic-predominant AD. In hippocampal-sparing AD, NFTs are increased in the cortex but relatively less so in the hippocampus (27). However, in limbic-predominant AD, NFT counts are normally increased in the hippocampus and decreased in the cortex. Meanwhile, typical AD usually has classic distributions of NFTs (28). According to neuroimaging patterns of CSF biomarkers, cognitive and clinical measures, white matter hyperintensities, and longitudinal trajectories, AD can be divided into four atrophy patterns: (I) patients with generally standard neuroanatomical profiles with slightly irregular cognitive and CSF biomarker profiles and the slowest clinical development; (II) patients with classical AD neuroanatomical, cognitive, and CSF biomarkers with a clinical profile displaying the quickest clinical development; (III) patients with a diffused array of deterioration with comparatively less distinct participation of the medial temporal lobe, irregular CSF amyloid-b1-42 values, and proportionately superior executive damage; (IV) patients with remarkably central participation of the medial temporal lobe and a slow stable development, possibly demonstrating in early AD stages (5). These biomarkers for staging AD offer the prospect for allowing accurate diagnosis, prognosis, and precise patient enrolment of comparatively consistent subgroups of individuals for clinical trials.
In this study, the GSE1297 and GSE28146 datasets, which focused on the critical early stages, were chosen for further study. GSE1297 dataset was utilized for AD-related modules and gene selection. Another dataset was used for independent verification. After some processes such as standardization and normalization, 9,974 genes were finally used for WGCNA. Then, 16 co-expression modules were recognized by the dynamic tree cut method. Through associating gene modules with three clinical features, the pink module was identified as having the greatest positive association with the pathological stage. Exploring the association between the pink module and pathological stage could narrow the search for those genes with vital biological importance to AD. There were six hub genes in the pink module, including AACS, GRIK1, HOXB2, KCNF1, MYBL1, and RAP1GAP2. Of these hub genes, mutations in the PSEN1 gene have been found to be linked with familial AD (29). The transcriptional factor HOXB2 has been proven to be up-regulated with folic acid to augment the effectiveness of memantine for spatial learning and neuronal defense (30). MYBL1 has shown different patterns in males’ and females’ molecular networks of AD based on global gene expression and integrative network analyses (31). Bos taurus RAP1GAP2 acts together with synaptotagmin-like protein 1 and Rab27 to control the excretion of dense granules from platelets at locations of endothelial injury, which may impact the progress of AD (32). KCNF1 has diverse functions, including regulating neurotransmitter release and AACS-related pathways of metabolism and butanoate metabolism, which may play roles in AD (33).
GRIK1, also known as glutamate receptor 5 (GluR5), is one of 14 receptor subunits. The glutamate receptor (GluR) subunits are included in ionotropic GluRs and regulate excitatory neurotransmission (34). A physical map of the GRIK1 region and a schematic representation of the splicing mechanisms were presented by Barbon et al. in 2000 (35). In 2014, Kranzler et al. showed that GRIK1 could reduce the heavy drinking in problem drinkers and that it might be a moderator for heavy drinkers (36). The function of GRIK1 in the central nervous system has not been fully characterized. GRIK1 was shown to be overexpressed in senile plaques (in the temporal lobe) in RNA-sequencing data of AD patients and may play a critical role in AD (37). However, the relationship between AD and GRIK1 is poorly understood. Therefore, we next investigated the level of GRIK1 in a mouse model. The results showed that the expression of GRIK1 was upregulated in both the cortex and hippocampus in AD mice, indicating a potential role in the pathogenesis of AD.
Enrichment analyses for the pink module specified that the biological processes of the pink module concentrated on PI3K signaling and ion transmembrane transport. Previous studies have suggested that the PI3K/Akt cascade is involved in the nicotine-induced neuroprotection through the phosphorylation of bad, a Bcl-2 family protein supporting neuronal endurance (38). Kihara et al. reported that nicotine treatment could promote the phosphorylation of Akt and the manifestation of Bcl-2 while protecting neurons from b-amyloid-enhanced glutamate neurotoxicity. Treatment with PI3K inhibitors like LY294002 and wortmannin can suppress the nicotine-induced neuroprotective impact, indicating that the PI3K/Akt cascade regulates the nicotine-induced neuroprotection through the Bcl-2 protein (39). Donepezil, galantamine, tacrine, specified as “therapeutic AChE inhibitors” and used in the treatment of AD, were shown to protect the principal cultures of rat cortical neurons from glutamate-induced neurotoxicity that includes apoptotic neuronal death via PI3K/Akt pathway (40).
One of the pathological changes in AD is the irregular proteolytic handling of APP, which is the vital step that causes the creation of amyloid plaque, NFTs, and neuronal loss. By interacting with ion transport systems, amyloid protein induces an array of effects via the activation of several mechanisms (41). Impairment of Ca2+ homeostasis, along with the production of reactive oxygen species was considered as the cause of amyloid protein-prompted cytotoxicity. The change in Ca2+ homeostasis may change endogenous ion transport systems, like as Ca2+ and K+ channels, ligand-driven ion channels, and G-protein-induced releases of second messengers or form heterogeneous ion channels. Amyloid proteins in AD modify numerous ion transport systems and form heterogeneous ion channels, which can alter cell regulation (42). These modifications harm the membrane by lowering its reliability and increasing its ion penetrability, thus perhaps promoting the progress of AD (42).
Moreover, we found that genes in the pink module can be used to discriminate between three kinds of AD samples of the incipient, moderate, and severe type, particularly between severe and non-severe AD samples in the GSE28146 validation set. Furthermore, ROC curve analysis was applied to assess the diagnostic effectiveness of six hub genes to discriminate between severe and non-severe AD samples. We found that the AUC value was higher than 0.7, but that the AUC values for less related modules (modules of lightcyan, salmon, and blue) were lower than 0.7, indicating that these six hub genes are potential biomarkers of AD stages.
In conclusion, we employed WGCNA to systematically recognize co-expressed gene modules and those hub genes related to AD development. One gene module and six hub genes capable of distinguishing between AD stages were identified. These outcomes can contribute to an improved understanding of the mechanisms participating in the development of AD.
Acknowledgments
None.
Footnote
Conflict of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Animal experiments were conducted in accordance with the guideline for the Care and Use of Laboratory Animals published by the US National Institutes of Health. The study was approved by the Ethics Committee of First Affiliated Hospital of Harbin Medical University, China (No. 2019022).
References
- Albert MS, DeKosky ST, Dickson D, et al. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 2011;7:270-9. [Crossref] [PubMed]
- Vemuri P, Lesnick TG, Przybelski SA, et al. Effect of lifestyle activities on Alzheimer disease biomarkers and cognition. Ann Neurol 2012;72:730-8. [Crossref] [PubMed]
- Sperling RA, Aisen PS, Beckett LA, et al. Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 2011;7:280-92. [Crossref] [PubMed]
- Mielke MM, Vemuri P, Rocca WA. Clinical epidemiology of Alzheimer's disease: assessing sex and gender differences. Clin Epidemiol 2014;6:37-48. [Crossref] [PubMed]
- Braak H, Braak E. Staging of Alzheimer's disease-related neurofibrillary changes. Neurobiol Aging 1995;16:271-8; discussion 278-84. [Crossref] [PubMed]
- Galasko D, Klauber MR, Hofstetter CR, et al. The Mini-Mental State Examination in the early diagnosis of Alzheimer's disease. Arch Neurol 1990;47:49-52. [Crossref] [PubMed]
- Bierer LM, Hof PR, Purohit DP, et al. Neocortical neurofibrillary tangles correlate with dementia severity in Alzheimer's disease. Arch Neurol 1995;52:81-8. [Crossref] [PubMed]
- Wang YQ, Qu DH, Wang K. Therapeutic approaches to Alzheimer's disease through stimulating of non-amyloidogenic processing of amyloid precursor protein. Eur Rev Med Pharmacol Sci 2016;20:2389-403. [PubMed]
- Blennow K, Hampel H, Weiner M, et al. Cerebrospinal fluid and plasma biomarkers in Alzheimer disease. Nat Rev Neurol 2010;6:131-44. [Crossref] [PubMed]
- Fagan AM, Mintun MA, Mach RH, et al. Inverse relation between in vivo amyloid imaging load and cerebrospinal fluid Abeta42 in humans. Ann Neurol 2006;59:512-9. [Crossref] [PubMed]
- Forsberg A, Engler H, Almkvist O, et al. PET imaging of amyloid deposition in patients with mild cognitive impairment. Neurobiol Aging 2008;29:1456-65. [Crossref] [PubMed]
- Hesse C, Rosengren L, Andreasen N, et al. Transient increase in total tau but not phospho-tau in human cerebrospinal fluid after acute stroke. Neurosci Lett 2001;297:187-90. [Crossref] [PubMed]
- Ost M, Nylen K, Csajbok L, et al. Initial CSF total tau correlates with 1-year outcome in patients with traumatic brain injury. Neurology 2006;67:1600-4. [Crossref] [PubMed]
- Hampel H, Burger K, Pruessner JC, et al. Correlation of cerebrospinal fluid levels of tau protein phosphorylated at threonine 231 with rates of hippocampal atrophy in Alzheimer disease. Arch Neurol 2005;62:770-3. [Crossref] [PubMed]
- Buerger K, Ewers M, Pirttila T, et al. CSF phosphorylated tau protein correlates with neocortical neurofibrillary pathology in Alzheimer's disease. Brain 2006;129:3035-41. [Crossref] [PubMed]
- Villemagne VL, Pike KE, Chetelat G, et al. Longitudinal assessment of Abeta and cognition in aging and Alzheimer disease. Ann Neurol 2011;69:181-92. [Crossref] [PubMed]
- Carlson MR, Zhang B, Fang Z, et al. Gene connectivity, function, and sequence conservation: predictions from modular yeast co-expression networks. BMC Genomics 2006;7:40. [Crossref] [PubMed]
- Zhou XG, Huang XL, Liang SY, et al. Identifying miRNA and gene modules of colon cancer associated with pathological stage by weighted gene co-expression network analysis. Onco Targets Ther 2018;11:2815-30. [Crossref] [PubMed]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [Crossref] [PubMed]
- Liu J, Zhou S, Li S, et al. Eleven genes associated with progression and prognosis of endometrial cancer (EC) identified by comprehensive bioinformatics analysis. Cancer Cell Int 2019;19:136. [Crossref] [PubMed]
- Zhao B, Chen Y, Hu S, et al. Systematic Analysis of Non-coding RNAs Involved in the Angora Rabbit (Oryctolagus cuniculus) Hair Follicle Cycle by RNA Sequencing. Front Genet 2019;10:407. [Crossref] [PubMed]
- Parsana P, Ruberman C, Jaffe AE, et al. Addressing confounding artifacts in reconstruction of gene co-expression networks. Genome Biol 2019;20:94. [Crossref] [PubMed]
- Blalock EM, Geddes JW, Chen KC, et al. Incipient Alzheimer's disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci U S A 2004;101:2173-8. [Crossref] [PubMed]
- Blalock EM, Buechel HM, Popovic J, et al. Microarray analyses of laser-captured hippocampus reveal distinct gray and white matter signatures associated with incipient Alzheimer's disease. J Chem Neuroanat 2011;42:118-26. [Crossref] [PubMed]
- Huang DW, Sherman BT, Tan Q, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 2007;35:W169-75. [Crossref] [PubMed]
- Si Z, Wang X, Zhang Z, et al. Heme Oxygenase 1 Induces Tau Oligomer Formation and Synapse Aberrations in Hippocampal Neurons. J Alzheimers Dis 2018;65:409-19. [Crossref] [PubMed]
- Murray ME, Graff-Radford NR, Ross OA, et al. Neuropathologically defined subtypes of Alzheimer's disease with distinct clinical characteristics: a retrospective study. Lancet Neurol 2011;10:785-96. [Crossref] [PubMed]
- Whitwell JL, Dickson DW, Murray ME, et al. Neuroimaging correlates of pathologically defined subtypes of Alzheimer's disease: a case-control study. Lancet Neurol 2012;11:868-77. [Crossref] [PubMed]
- Antonell A, Llado A, Altirriba J, et al. A preliminary study of the whole-genome expression profile of sporadic and monogenic early-onset Alzheimer's disease. Neurobiol Aging 2013;34:1772-8. [Crossref] [PubMed]
- Chen TF, Huang RF, Lin SE, et al. Folic Acid potentiates the effect of memantine on spatial learning and neuronal protection in an Alzheimer's disease transgenic model. J Alzheimers Dis 2010;20:607-15. [Crossref] [PubMed]
- Sun LL, Yang SL, Sun H, et al. Molecular differences in Alzheimer's disease between male and female patients determined by integrative network analysis. J Cell Mol Med 2019;23:47-58. [Crossref] [PubMed]
- Willard FS, Low AB, McCudden CR, et al. Differential G-alpha interaction capacities of the GoLoco motifs in Rap GTPase activating proteins. Cell Signal 2007;19:428-38. [Crossref] [PubMed]
- Gutman GA, Chandy KG, Grissmer S, et al. International Union of Pharmacology. LIII. Nomenclature and molecular relationships of voltage-gated potassium channels. Pharmacol Rev 2005;57:473-508. [Crossref] [PubMed]
- Sommer B, Seeburg PH. Glutamate receptor channels: novel properties and new clones. Trends Pharmacol Sci 1992;13:291-6. [Crossref] [PubMed]
- Barbon A, Barlati S. Genomic organization, proposed alternative splicing mechanisms, and RNA editing structure of GRIK1. Cytogenet Cell Genet 2000;88:236-9. [Crossref] [PubMed]
- Kranzler HR, Covault J, Feinn R, et al. Topiramate treatment for heavy drinkers: moderation by a GRIK1 polymorphism. Am J Psychiatry 2014;171:445-52. [Crossref] [PubMed]
- Twine NA, Janitz K, Wilkins MR, et al. Whole transcriptome sequencing reveals gene expression and splicing differences in brain regions affected by Alzheimer's disease. PLoS One 2011;6:e16266. [Crossref] [PubMed]
- del Peso L, Gonzalez-Garcia M, Page C, et al. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science 1997;278:687-9. [Crossref] [PubMed]
- Kihara T, Shimohama S, Sawada H, et al. alpha 7 nicotinic receptor transduces signals to phosphatidylinositol 3-kinase to block A beta-amyloid-induced neurotoxicity. J Biol Chem 2001;276:13541-6. [Crossref] [PubMed]
- Takada-Takatori Y, Kume T, Sugimoto M, et al. Acetylcholinesterase inhibitors used in treatment of Alzheimer's disease prevent glutamate neurotoxicity via nicotinic acetylcholine receptors and phosphatidylinositol 3-kinase cascade. Neuropharmacology 2006;51:474-86. [Crossref] [PubMed]
- Sultana R, Newman SF, Abdul HM, et al. Protective effect of D609 against amyloid-beta1-42-induced oxidative modification of neuronal proteins: redox proteomics study. J Neurosci Res 2006;84:409-17. [Crossref] [PubMed]
- Casey NH, Crosley RI, Smith GA. Influence of continuous dietary halofuginone on broiler skin tensile strength and growth performance. J S Afr Vet Assoc 1992;63:16-9. [PubMed]


