
Cryo-electron microscopy reveals informative details of GABAA receptor structural pharmacology: implications for drug discovery
The first cryoEM high-resolution structure of a recombinant full-length heterotrimeric γ-aminobutyric acid (GABA) type A receptor (GABAR) subtype α1β3γ2L in complex with important GABAR ligands was published by the Aricescu lab in January 2019 (1,2). GABARs are the major receptors mediating rapid inhibitory neurotransmission in the central nervous system and members of the pentameric ligand-gated ion channel (pLGIC) gene superfamily. The α1β3γ2L recombinant GABAR was isolated from a stable cell line and reconstituted into a lipid bilayer. It has a megabody (Mb38) attached that binds with high affinity to the α1+/β3- subunit interface and acts as a positive allosteric modulator (PAM) (2). Five other structures with additional ligands bound were solved: picrotoxin (PTX, being an open channel pore blocker) alone and PTX with GABA; bicuculline, a competitive antagonist of the GABA site, and two positive allosteric modulatory benzodiazepine (BZ) drugs, diazepam and alprazolam (1). The two α1β3γ2L GABAR cryoEM papers provide exquisite detail for ligand binding sites for agonists/antagonists and several kinds of positive allosteric modulators (PAMs) binding both to the extracellular domain (ECD) (i.e., GABA, BZs) and the trans-membrane domain (TMD) Cl- channel pore (PTX). The details confirm and explain previous studies and structural modeling of GABARs, yet the high resolution provides a significantly greater understanding of agonist (GABA) ligand binding at 2 of the 5 different subunit interfaces in the ECD, and channel gating involving cross-talk of the agonist-bound ECD stabilizing allosterically the open state of the TMD channel. It provides details on how PTX blocks the pore and at the same time allosterically modulates ligand binding in GABARs (1). Furthermore, molecular mechanisms are revealed for PAMs, the benzodiazepines, that bind at modified GABA sites at a third subunit interface in the ECD, showing structural explanations for GABAR subtype specificity and efficacy of different ligand classes of BZ and non-BZ ligands for the BZ sites.
The purification of GABAR protein for cryoEM imaging did not require the amounts nor the purity of the protein needed for X-ray crystallography, but still required a large-scale production, provided by expression of epitope-tagged recombinant GABAR in a stably expressing and inducible human cell line (3). Others had succeeded in producing electron microscopy structures of pLGIC such as electric fish nicotinic acetylcholine receptors (4) and X-ray crystal structures of the GluCl protein from nematodes (5). However, the structures of mammalian GABARs had been homomeric models, including the homomeric, trimmed (to allow crystal formation) β3 GABAR X-ray structure (6) and chimeric structures (7). Dramatically improved technology for single-particle electron cryomicroscopy (cryoEM) data collection and digital processing, model building and refinement, have produced high-resolution structures of membrane proteins including pLGIC (8). The first cryoEM structures of heterodimeric [α5β3: (9)] and heterotrimeric (α1β2γ2) (10,11) GABARs appeared in 2018. These last two breakthrough papers, while lauded, were immediately also criticized [e.g., Sigel, 2018 (12)] for imperfections, with possible problems in resolution of the TMD showing a collapsed pore wall at the γ2 subunit. The main problem was suggested to be due to detergent damage, as supported by the Aricescu group’s results discussed here (1,2). This was solved with the reconstitution in phospholipid bilayers (nanodiscs) as pioneered earlier (13,14). An additional improvement in these two recent papers was to utilize full-length sequences for all three subunits, including the intracellular domains (ICD), although the large M3-M4 loop was not resolved, likely because it is largely unstructured in recombinantly expressed α1β3γ2 GABARs. The ICDs are likely needed for interactions with modulatory proteins in the neurons, and maybe also in heterologous cells. We will return to this topic later when discussing the vagaries of utilizing heterologous cell expression of recombinant proteins as opposed to native tissue sources.
GABA binding to the two ECD β3+/α- subunit interfaces (Figure 1) is consistent with models based on previous work, yet show in detail how GABA binding leads to structural changes resulting in receptor activation. This includes (I) stabilizing the counterclockwise twist of the ECD, producing channel gating in the TMD (see Figure 2A) and, (II) providing a detailed molecular understanding of the Monod-Wyman-Changeux (MWC) allosteric model (15) demonstrated for membrane proteins with pLGIC by Changeux (16).
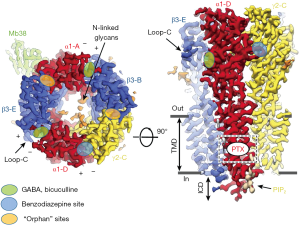
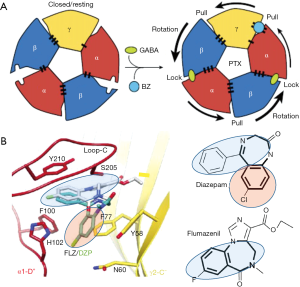
Arrangement of subunits (top or bottom view, Figure 1) fits canonical models based on earlier work, but with new designation of ABCDE for αβγαβ (clockwise, viewed from extracellular space). Two β+/α- interface GABA-bicuculline sites of course differ from the other 3 interfaces, but also from each other with differences in binding affinity (17). It is remarkable that there are indeed structural differences discernible in the cryoEM structure distinguishing the β3-B+/α1-A– from the β3-E+/α1-D– GABA binding site (1).
Other important details that emerged from these studies are differences in binding modes of the classical BZs (diazepam and alprazolam) versus the imidazobenzodiazepines (iBZs) (flumazenil, bretazenil) (see Figure 2B). The classical BZs diazepam and alprazolam are clinically used as sedative, anti-anxiety, and anti-convulsant drugs, whereas flumazenil is the widely used BZ-site antagonist used to treat BZ-site drug overdoses and to reverse BZ actions after clinical procedures. Interestingly the BZ core structures (bold in Figure 2B) for BZs and iBZs show different orientations in the cryoEM structures, which may explain their different pharmacology on GABAR subtypes, with α4 and α6 subunit-containing receptors having low affinity for classical BZ like diazepam and alprazolam, but retaining their high affinity for iBZs like flumazenil and its close relative Ro15-4513. Details on how BZ-site drugs (which includes non-BZs like zolpidem) fit into their binding site hopefully will give new impetus to find e.g., specific anxiolytic BZ-site drugs that lack sedative actions (18-20).
Another area discussed by the two 2019 Aricescu papers (1,2) was the use of the cryoEM structures of the GABAR TMD to confirm and extend models based on earlier studies on general anesthetic PAM ligand sites (intravenous agents: barbiturates, etomidate, propofol, neuroactive steroids; volatile agents; and alcohols, including high concentrations (~100 mM) of ethanol. These anesthetic sites are defined by mutations in TMD pore residues that eliminate or reduce anesthetic sensitivity in vitro (21) and in β3-N265M knock-in mice in vivo (22). Affinity labeling and sequencing demonstrated binding and PAM action of anesthetics on the outer side of the TMD helical residues (23). Like those for GABA and BZ in the ECD, the evidence suggests that these binding sites are mostly at subunit interfaces. Similar domains were employed in the TMD of the five different interfaces, but, not surprisingly, the selectivity varied for different chemical classes of PAMs (24). Future GABAR cryoEM structures with these anesthetics will help to explain their selectivity, and reveal mechanistic insights.
Masiulis et al. (1) showed that the endogenous phospholipid PIP2 (phosphatidylinositol-4,5-bisphosphate) is a natural structural, potentially modulatory feature of GABARs, with two PIP2 molecules at the periphery of the two α1 subunits in the α1β3γ2 receptor. Positively charged cytoplasmic peri-TMD region α1 subunit residues bind negatively charged PIP2 phosphate groups. These charged peri-TMD PIP2 binding residues are identical in α1,2,3,5 subunits, but not in α4 and α6 subunits (2), raising the interesting possibility that α4 and α6 subunit-containing GABARs differ in terms of PIP2 binding/modulation. Note that GABARs α4 and α6 subunits, when partnered with the δ subunit, form pharmacologically and physiologically highly distinct extrasynaptic GABARs (25-27). Laverty et al. (2) suggested that PIP2 might directly regulate GABAR channel gating as seen with several other ion channels [for review see Hille et al. (28)]. Particularly noteworthy are two studies on the structural basis of PIP2 activation of (I) the Kir2.2 inwardly rectifying K+ channel (29) and (II) the TRPV1 via PIP2 binding to the capsaicin binding site (13). GABAR synaptic clustering involving the matrix protein gephyrin has been shown to be modulated by PIPx (30). Almost certainly there will be future studies to investigate the potential role of PIP2, e.g., in the modulation of synaptic GABAR plasticity and/or receptor trafficking and possibly also subunit assembly.
While these cryoEM studies on α1β3γ2 receptors assembled into the lipid environment of nanodiscs provide a number of truly amazing breakthroughs, recombinantly expressed GABAR may lack endogenous assembly, trafficking and clustering proteins, auxiliary subunits, lipids, and post-translational modifications (glycosylation, phosphorylation, methylation, etc.), and may even assemble into a non-native architecture. It will be therefore important to study native brain GABAR proteins purified from mammalian brains. This was recently achieved by the Gouaux group who solved the cryoEM structures of a whole family of native brain AMPAR subtype of excitatory glutamate receptor LGICs, with dramatically new refined structural information not previously demonstrated by structural work on recombinant glutamate LGIC receptors (31).
Together, these reports provide a tremendous potential for structure-based drug discovery of better therapeutic agents for the myriad neuropsychiatric disorders treatable with GABAR drugs (19,20,32,33) with improved selectivity and reduced side effects. Visualizing the binding of small molecule ligands at the low Angstrom resolution gives new impact to quantitative structure-function activity relationships. Coupled with a greater understanding emerging recently for GABAR subtype selectivity of PAM action using genetically engineered mice (18) or affinity labeling and site-directed mutagenesis for verification of PAM sites (24) and of brain circuitry roles in the clinical disorders using cell type/brain region-selective optogenetic knockdown with viral vectors in vivo (34), exciting possibilities appear almost certainly forthcoming with these breakthroughs in structural pharmacology.
Other topics briefly addressed by the two Aricescu papers (including methods and extended data on line) include translational aspects of normal and diseased brain. For example, Laverty et al., 2019 examined the structural effects of mutations in GABAR producing human diseases (extended data figure 7), such as epilepsy. Epileptologists doing structure-function studies on these mutations in vitro and in knock-in rodents are already applying the new structural information to new experiments (35). The importance of cryoEM to explain complex cellular and brain functions, diseases, and structure-based drug design cannot be overstated.
These findings and insights provide inspiration and useful clues for studying a wealth of other questions about GABARs including physiologically and pharmacologically unique extrasynaptic δ-GABAR. These extrasynaptic receptors show high sensitivity to GABA [and GABA analogs like muscimol and THIP/Gaboxadol (25,27)]. While δ-GABARs and extrasynaptic GABA currents are insensitive to classical BZs, they are highly sensitive to alcohol (26) with a proposed ethanol binding site at the ECD (36). The ECD α+/β- interface binds the megabody protein Mb38 (see Figure 1, left) (1) and this site has been recently shown to be an allosteric modulatory site for selected ligands (19,37).
Acknowledgments
Funding: The authors were supported by NIH grant AA021213 to RW Olsen and M Wallner.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Masiulis S, Desai R, Uchanski T, et al. GABAA receptor signalling mechanisms revealed by structural pharmacology. Nature 2019;565:454-9. [Crossref] [PubMed]
- Laverty D, Desai R, Uchanski T, et al. Cryo-EM structure of the human α1β3γ2 GABAA receptor in a lipid bilayer. Nature 2019;565:516-20. [Crossref] [PubMed]
- Dostalova Z, Zhou X, Liu A, et al. Human α1β3γ2L GABAA receptors: High-level production and purification in a functional state. Protein Sci 2014;23:157-66. [Crossref] [PubMed]
- Unwin N. Nicotinic acetylcholine receptor and the structural basis of neuromuscular transmission: insights from Torpedo postsynaptic membranes. Q Rev Biophys 2013;46:283-322. [Crossref] [PubMed]
- Hibbs RE, Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 2011;474:54-60. [Crossref] [PubMed]
- Miller PS, Aricescu AR. Crystal structure of a human GABA receptor. Nature 2014;512:270-5. [Crossref] [PubMed]
- Laverty D, Thomas P, Field M, et al. Crystal structures of a GABAA-receptor chimera reveal new endogenous neurosteroid-binding sites. Nat Struct Mol Biol 2017;24:977-85. [Crossref] [PubMed]
- Cressey D, Callaway E. Cryo-electron microscopy wins chemistry Nobel. Nature 2017;550:167. [Crossref] [PubMed]
- Liu S, Xu L, Guan F, et al. Cryo-EM structure of the human α5β3 GABAA receptor. Cell Res 2018;28:958-61. [Crossref] [PubMed]
- Phulera S, Zhu H, Yu J, et al. Cryo-EM structure of the benzodiazepine-sensitive α1β1γ2S tri-heteromeric GABAA receptor in complex with GABA. Elife 2018.7. [PubMed]
- Zhu S, Noviello CM, Teng J, et al. Structure of a human synaptic GABAA receptor. Nature 2018;559:67-72. [Crossref] [PubMed]
- Sigel E. Key receptor involved in neuronal signalling visualized. Nature 2018;559:37-8. [Crossref] [PubMed]
- Gao Y, Cao E, Julius D, et al. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 2016;534:347-51. [Crossref] [PubMed]
- Denisov IG, Sligar SG. Nanodiscs for structural and functional studies of membrane proteins. Nat Struct Mol Biol 2016;23:481-6. [Crossref] [PubMed]
- Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: a plausible model. J Mol Biol 1965;12:88-118. [Crossref] [PubMed]
- Changeux JP, Christopoulos A. Allosteric modulation as a unifying mechanism for receptor function and regulation. Cell 2016;166:1084-102. [Crossref] [PubMed]
- Baumann SW, Baur R, Sigel E. Individual properties of the two functional agonist sites in GABAA receptors. J Neurosci 2003;23:11158-66. [Crossref] [PubMed]
- Möhler H, Fritschy JM, Rudolph U. A new benzodiazepine pharmacology. J Pharmacol Exp Ther 2002;300:2-8. [Crossref] [PubMed]
- Sigel E, Ernst M. The benzodiazepine binding sites of GABAA receptors. Trends Pharmacol Sci 2018;39:659-71. [Crossref] [PubMed]
- Solomon VR, Tallapragada VJ, Chebib M, et al. GABA allosteric modulators: An overview of recent developments in non-benzodiazepine modulators. Eur J Med Chem 2019;171:434-61. [Crossref] [PubMed]
- Mihic SJ, Ye Q, Wick MJ, et al. Sites of alcohol and volatile anaesthetic action on GABAA and glycine receptors. Nature 1997;389:385-9. [Crossref] [PubMed]
- Jurd R, Arras M, Lambert S, et al. General anesthetic actions in vivo strongly attenuated by a point mutation in the GABAA receptor β3 subunit. Faseb J 2003;17:250-2. [Crossref] [PubMed]
- Li GD, Chiara DC, Sawyer GW, et al. Identification of a GABAA receptor anesthetic binding site at subunit interfaces by photolabeling with an etomidate analog. J Neurosci 2006;26:11599-605. [Crossref] [PubMed]
- Forman SA, Miller KW. Mapping general anesthetic sites in heteromeric GABAA receptors reveals a potential for targeting receptor subtypes. Anesth Analg 2016;123:1263-73. [Crossref] [PubMed]
- Benkherouf AY, Taina KR, Meera P, et al. Extrasynaptic δ-GABAA receptors are high-affinity muscimol receptors. J Neurochem 2019;149:41-53. [Crossref] [PubMed]
- Hanchar HJ, Dodson PD, Olsen RW, et al. Alcohol-induced motor impairment caused by increased extrasynaptic GABAA receptor activity. Nat Neurosci 2005;8:339-45. [Crossref] [PubMed]
- Meera P, Wallner M, Otis TS. Molecular basis for the high THIP/gaboxadol sensitivity of extrasynaptic GABAA receptors. J Neurophysiol 2011;106:2057-64. [Crossref] [PubMed]
- Hille B, Dickson EJ, Kruse M, et al. Phosphoinositides regulate ion channels. Biochim Biophys Acta 2015;1851:844-56. [Crossref] [PubMed]
- Hansen SB, Tao X, MacKinnon R. Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature 2011;477:495-8. [Crossref] [PubMed]
- Papadopoulos T, Rhee HJ, Subramanian D, et al. Endosomal Phosphatidylinositol 3-Phosphate Promotes Gephyrin Clustering and GABAergic Neurotransmission at Inhibitory Postsynapses. J Biol Chem 2017;292:1160-77. [Crossref] [PubMed]
- Zhao Y, Chen S, Swensen AC, et al. Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM. Science 2019;364:355-62. [Crossref] [PubMed]
- Olsen RW, Sieghart W. International Union of Pharmacology. LXX. Subtypes of GABAA receptors: classification on the basis of subunit composition, pharmacology, and function. Update. Pharmacol Rev 2008;60:243-60. [Crossref] [PubMed]
- Olsen RW. GABAA receptor: Positive and negative allosteric modulators. Neuropharmacology 2018;136:10-22. [Crossref] [PubMed]
- Engin E, Benham RS, Rudolph U. An emerging circuit pharmacology of GABAA receptors. Trends Pharmacol Sci 2018;39:710-32. [Crossref] [PubMed]
- Hernandez CC. Altered inhibitory synapses in de novo GABRA5 and GABRA1 mutations associated with early onset epileptic encephalopathies. Brain 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Wallner M, Hanchar HJ, Olsen RW. Alcohol selectivity of β3-containing GABAA receptors: Evidence for a unique extracellular alcohol/imidazobenzodiazepine Ro15-4513 binding site at the α+β- subunit interface in αβ3δ GABAA receptors. Neurochem Res 2014;39:1118-26. [Crossref] [PubMed]
- Ramerstorfer J, Furtmuller R, Sarto-Jackson I, et al. The GABAA receptor α+β- interface: a novel target for subtype selective drugs. J Neurosci 2011;31:870-7. [Crossref] [PubMed]


