
Differentially expressed liver exosome-related genes as candidate prognostic biomarkers for hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) accounts for 85–90% of primary liver cancers and is the fourth leading cause of cancer-related death worldwide. Approximately 841,000 new HCC cases are diagnosed worldwide each year (1,2). Despite new breakthroughs in targeted therapy, immune therapy, interventional therapy, surgical techniques, and liver transplantation, the prognosis of HCC remains poor due to its high rates of metastasis and recurrence (3-6). Traditional prognostic models for HCC use clinic-pathological parameters, such as stage, and other parameters; however, due to tumor heterogeneity among patients, these models no longer work satisfactorily. Therefore, it is necessary to investigate the underlying molecular mechanisms of HCC and to identify new diagnostic and prognostic markers for the disease (7,8).
Exosomes are tiny extracellular vesicles with a lipid bilayer membrane structure that were first discovered by Johnstone in 1989 in his study of reticulocytes (9). Later studies revealed that exosomes can transport biologically active molecules between cells to regulate the microenvironment between cells and the immune system, and are closely related to tumorigenesis and tumor development (3,10,11). Exosomes derived from tumor tissue contain numerous cancer-related biomarkers, such as microRNAs (miRNAs), which can be used to detect HCC at an early stage. Although some breakthroughs have been made in the field of exosome research, the specific biological functions of exosomes have yet to be fully elucidated (12-15). Therefore, the aim of this study was to comprehensively investigate exosome-related genes in HCC, which can help predict the outcome of HCC and discover potential new drug candidates for specific targeted therapy. We present the following article in accordance with the TRIPOD reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-4400/rc).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Data collection
Genes related to liver-derived exosomes were downloaded from the exoRBase (http://www.exorbase.org/). Data on mRNA expression in 407 tumor and 58 normal tissue samples, and the corresponding clinical data, were obtained from The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/). For the validation dataset, data on gene expression in 240 tumor and 202 normal tissue samples were obtained from the International Cancer Genome Consortium (ICGC) database (https://dcc.icgc.org/). The study flowchart is shown in Figure 1.
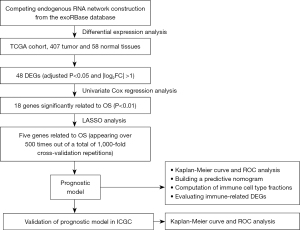
Identification of differentially expressed genes and competing endogenous RNA network construction
Differentially expressed genes (DEGs) related to exosomes and liver cancer were identified using the limma package in R software. Those DEGs with an adjusted P<0.05 and |log2(fold change)| >1 were selected for further analysis, as described below:
- The miRcode database (http://mircode.org/) was used to search for interactions between long non-coding RNAs (lncRNAs) and miRNAs, and to match miRNA interactions;
- The TargetScan (http://targetscan.org/) and miRanda (http://www.microrna.org/) databases were used to predict miRNA-targeted mRNAs;
- The Encyclopedia of RNA Interactomes database (http://starbase.sysu.edu.cn/) was used to predict the relationships between circular RNAs (circRNA) and miRNAs.
A competing endogenous RNA (ceRNA) network comprising mRNAs obtained from these databases that overlapped with the exosome-related mRNAs was constructed. Results were graphed by Cytoscape 3.7.2. [Cytoscape is provided by the U.S. National Institute of General Medical Sciences (NIGMS)].
Development and validation of a prognostic model
First, we performed univariate Cox regression analysis in TCGA database and two-sided P<0.05 was set as statistically significant. The glmnet package in R software was used to narrow the genetic screening range, after which least absolute shrinkage and selection operator (LASSO) regression was performed (16). We performed 1,000 replicates on the dataset and selected markers with >500 times replicates. The regression coefficient (β) was obtained from the LASSO regression. The prognostic index (PI) was then calculated using the following formula:
The X-tile software was used to establish the optimal cut-off value to divide patients into high- and low-risk groups. The predictive power of the prognostic model was evaluated using Kaplan-Meier survival analysis, and the model was validated in the ICGC database.
Identification of independent prognostic parameters for HCC
Independent factors affecting clinical prognosis of patients with HCC were obtained by univariate and multivariate Cox proportional hazards regression analyses. Two-sided P<0.05 was considered significant. Hazard ratios (HRs) and 95% confidence intervals (CIs) were calculated for each factor.
Establishment and evaluation of a forecast nomogram
Independent prognostic factors obtained from the Cox analyses were used to construct a nomogram to predict the 1- , 2- and 3-year survival rates of patients with HCC. The nomogram was verified internally using a calibration chart, and its predictive ability was assessed through time-varying receiver operating characteristic (ROC) curve analysis (17,18).
Immune cell type scores and immunotherapy effect
CIBERSORT is a new analytical tool to characterize cell composition based on gene expression profiling which is frequently used to evaluate immune cell infiltration (19). In the present study, CIBERSORT was used to forecast differences in the ratios of immune cell types between the low- and high-risk patient groups. The Genefilter package of R was used to screen each sample, and P<0.05 was considered significant.
Immunotherapy is a very promising treatment method for patients with HCC, and its therapeutic effect is related to molecules including programmed death ligand 1 (PD-L1), programmed death ligand 2 (PD-L2), T cell Ig and ITIM domain (TIGIT), and indoleamine-2,3-dioxygenase 1 (IDO1). Differences in the expression of these molecules between the low- and high-risk groups were evaluated.
Statistical analysis
Cox regression and LASSO regression analyses were used to develop the prognostic model, and univariate and multivariate Cox proportional hazards regression analyses were used to identify independent prognostic parameters for HCC. Two-tailed P<0.05 was considered significant. Statistical analyses were performed with R 4.0.2.
Results
Construction of the ceRNA network
Using data downloaded from the exoRBase database, differences in mRNA, lncRNA, and circRNA expression between tumor and non-tumor tissues were identified. Relationships between mRNAs, lncRNAs, miRNAs, and circRNAs, and between lncRNA-miRNA, miRNA-mRNA, and circRNA-miRNA pairs were explored. Then, miRNAs were used to develop a ceRNA network with Cytoscape software. In all, 722 lncRNA-miRNA pairs, 680 miRNA-mRNA pairs, and 64 circRNA-miRNA pairs were obtained. Finally, 19 differentially expressed (DE) lncRNAs, 45 DEmiRNAs, 47 DEmRNAs, and 2 DEcircRNAs were used to construct a ceRNA network.
Construction of an exosome-related gene prognostic model in TCGA
Based on intersections between mRNAs found in the ceRNA network and TCGA genes, 48 DEGs were identified. Univariate Cox regression analysis revealed 18 DEGs that were significantly correlated with overall survival (OS) in patients with HCC (P<0.01). LASSO analysis was used to narrow down the range of genes. Five DEGS that appeared >500 times in a total of 1,000 replicates were selected, and these genes (XPO1, IFI30, FBXO16, CALM1, MORC3) were chosen to establish a prognostic model using the following equation:
X-tile indicated that the best cut-off value to discriminate between high- and low-risk patients was 2.06. The patients were divided into low- and high-risk groups, and Kaplan-Meier analysis revealed that OS was significantly poorer for the high-risk group than the low-risk group (P<0.001; HR 3.51; 95% CI: 1.68–7.31; Figure 2A). The area under the time-dependent ROC curve for 3- and 5-year OS was 0.681 and 0.708, respectively, indicating the good predictive performance of the prognostic model (Figure 2B).
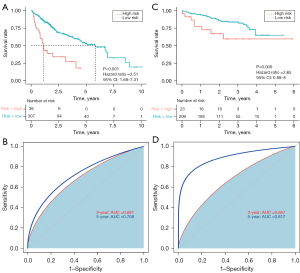
External validation of the exosome-related gene prognostic model in the ICGC
The predictive power of our prognostic model was validated using samples from the ICGC. As described above, patients were divided into low- and high-risk groups using a cut-off value of 2.06; the overall survival rate of patients in the low-risk group was higher than that of patients in the high-risk group (P=0.009; HR 2.65; 95% CI: 0.88–8; Figure 2C). The area under the curve of the five-gene prognostic model for 3- and 5-year survival was 0.667 and 0.817, respectively (Figure 2D), indicating the reliability of the model.
Interpretation of the model genes
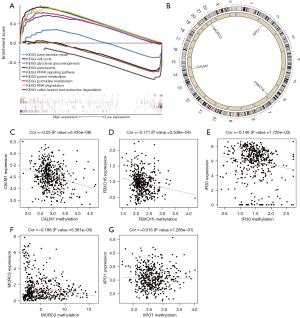
Gene set enrichment analysis functional annotation showed that the five HCC exosome-related genes in the prognostic model were enriched in the base excision repair, cell cycle, and glycolysis pathways (Figure 3A). As shown in Figure 3B, single deletions were found in XPO1 (2p15), FBXO16 (8p21.1), CALM1 (14q32.11), and MORC3 (21q22.13). Regression analysis between gene expression and methylation revealed that there were negative correlations between methylation and gene expression of CALM1, FBXO16, IFI30, and MORC3; however, no obvious correlation was found for XPO1 (Figure 3C-3G).
Establishing the predictive nomogram
Univariate Cox regression analysis revealed TNM stage and risk score to be significant predictors for survival (Figure 4A). Through multivariate Cox regression analysis, TNM stage and risk score were confirmed as independent prognostic factors (Figure 4B).
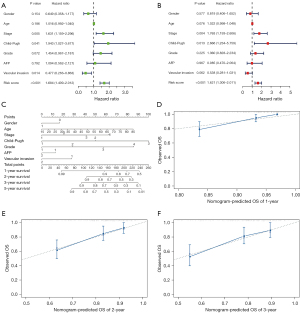
Subsequently, a nomogram including TNM stage and the risk score was built. Using the nomogram, scores are assigned to independent factors at each level for individual patients, with the total score calculated by summing these scores. By transforming the relationship of the total score, the 1-, 2- and 3-year survival rates can be obtained (Figure 4C). A calibration map was used for internal verification of the nomogram, which showed that there was good conformity between the predicted and observed results (Figure 4D-4F).
Computation of immune cell type fractions
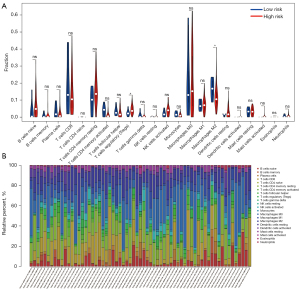
CIBERSORT was used to directly analyze the types and expression of immune cells in tissues from patients with HCC, combined with the Leukocyte signature matrix (LM22). Differences in 22 types of immune cells were examined between the low- and high-risk HCC groups. Significant differences were found for two types of immune cells: regulatory T (Treg) cells (P=0.043) and M2 macrophages (P=0.048; Figure 5A). Based on this result, we may conclude that high levels of Treg cells and M2 macrophages in patients with HCC are related to a poor prognosis. Figure 5B shows the proportion of each immune cell type in each sample, more intuitively showing the distribution of immune cells in different samples.
Evaluation of immune-related DEGs
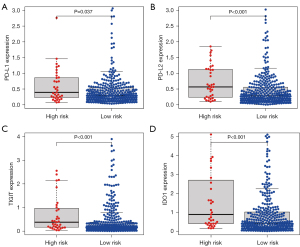
Blocking immune checkpoints is a promising method for treating many cancers. Therefore, in the present study we evaluated differences in the expression of key immune checkpoint molecules, including PD-L1, PD-L2, TIGIT, and IDO1, between low- and high-risk HCC patients. As shown in Figure 6A-6D, the expression levels of immune checkpoint molecules were higher in the high-risk group than in the low-risk group (P<0.05). Based on this finding, we may conclude that high expression levels of exosome-related genes are related to a poor prognosis of HCC, which may account for patients’ heterogeneous responses to immunotherapy and help to identify specific patient populations that may benefit from immunotherapy.
Discussion
Hepatocellular carcinoma is a fatal malignant tumor that has been a public health problem in many countries for many years. Although there has been considerable progress in the treatment of HCC, survival rates have not improved satisfactorily (19). Accumulating evidence shows that the malignant phenotype is affected by the cancer-related microenvironment. Hence, discovering immune biomarkers to predict the prognosis of patients with HCC is important and may also help with immunotherapy (20-22). In the present study, we used the TCGA database to develop a prognostic model based on exosome-related genes (XPO1, IFI30, FBXO16, CALM1, and MORC3) to predict the prognosis of patients with HCC, and then successfully validated the prediction ability of the model using an external database.
The genes used to build our prognostic model have been studied previously. Azizian et al. reported that XPO1 is frequently overexpressed in cancer cells and that its suppression leads to reductions in proteins such as MYC and epidermal growth factor receptor (20). XPO1 interacts with hundreds of proteins, which may affect its nuclear export activity. Originally identified as an autoantigen in inflammatory myopathies, MORC3 is recognized as a human ATPase and has been linked to cancer (21,22). CALM1 is a Ca2+ receptor protein which mediates a large number of signaling processes, including proliferation, motility, and differentiation. It had been found that CALM1 expression is strongly associated with many cancers (23). FBXO16 is a tumor suppressor that attenuates nuclear β‐catenin, and suppresses the growth, migration, and invasion of cancer cells (24). IFI30 has been found to be highly expressed in malignant glioma and to possibly have an influence on response to chemotherapy (25). Based on the results of the present study, IFI30 may affect the prognosis of HCC, but further studies are needed to illustrate the mechanism in detail. Furthermore, microenvironment analysis indicated that high IFI30 expression was accompanied by greater infiltration of M2 macrophages, which may be a research direction (25). Also, the five genes used in our prognostic model were found to be enriched mainly in the base excision repair, cell cycle, and glycolysis pathways, which have a clear relationship with tumor occurrence and development (26-29).
From another viewpoint, our model could be used to predict the prognosis of tumors and potential new treatment targets. Abnormal DNA methylation is well known to occur in all kinds of cancers and, in previous studies (30,31), we found that the expression of CALM1, FBXO16, IFI30, and MORC3 was negatively correlated with their methylation. Normal cells can transform to tumor cells through the occurrence of driver mutations and then the process of methylation; from this, it can be inferred that these genes play a role in tumorigenesis.
Liu et al. found that functional Treg cells facilitate the development of an M2 macrophage phenotype by repressing the CD8+ T cell-interferon γ axis and promoting the mitochondrial integrity of M2 macrophages via CD8+ T cells (32). Based on their study, it can be inferred that there is a positive correlation between Treg cells and M2 macrophages; however, in the present study, the opposite was observed, which may indicate that, in HCC, tumorigenesis and progression may occur as a result of changes in these pathways.
We confirmed that the prognostic model we developed can divide patients with HCC into low- and high-risk groups, and then showed that the oncologic outcomes of the low-risk group were better than those of the high-risk group. A nomogram was then built to predict the 1-, 2-, and 3-year survival rates. The calibration plot demonstrated that the resulting curve was very close to the real (observed data) curve, which means well predictive value. Thus, we further analyzed differences in infiltrating immune cells and immune-related gene expression between the low- and high-risk groups. In the present study, high-risk patients were more likely than low-risk patients to have higher levels of immune checkpoint molecules and a poor prognosis.
Many studies have focused on the role of exosomes in intercellular communication in HCC. It is widely accepted that HCC cells communicate with normal cells and promote the development and metastasis of HCC through exosomes. Some exosomes regulate the signal transduction pathway between cells, whereas others can be used as medicines due to the protective effects of the outer membrane (33,34). However, there is a lack of research on exosome-related genes for predicting survival and the effect of immunotherapy in HCC. In recent years, there have been breakthroughs in the treatment of liver cancer, especially in immunotherapy (35,36); the present study adds to the bank of exploratory research in this area and will hopefully contribute to future studies.
However, this study has some limitations. First, our study was retrospective and its findings need to be further verified in prospective studies, especially randomized control trials. Second, the LASSO analysis may have resulted in some important factors that contribute to the prognosis of HCC being overlooked. Third, the expression and the prognostic effects of the five genes used to develop the model require further investigation at the protein level. Fourth, functional experiments are needed to clarify it’s the potential mechanism. Finally, the detailed mechanism between tumor cells and immune cells requires further clarification.
Conclusions
This study has identified a novel signature based on exosome-related genes that has potential as a biomarker for the prognosis of HCC. These results may provide an immunological perspective for the development of precision treatment for HCC.
Acknowledgments
The authors acknowledge all those who have made a contribution to this article. The authors thank Jodi Cory Reed, PhD, from AJE Editing China (https://www.aje.cn/), for language English editing a draft of this manuscript.
Funding: This work was supported by the International Science and Technology Cooperation Projects (No. 2016YFE0107100), CAMS Clinical and Translational Medicine Research Funds (No. 2019XK320006), CAMS Innovation Fund for Medical Science (No. 2017-I2M-4-003 and 2018-I2M-3-001), Beijing Natural Science Foundation (Nos. L172055 and 7192158), the Fundamental Research Funds for the Central Universities (No. 3332018032), CSCO-Hengrui Cancer Research Fund (No. Y-HR2019-0239), and the National Ten-Thousand Talent Program.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-4400/rc
Peer Review File: Available at https://atm.amegroups.com/article/view/10.21037/atm-21-4400/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-4400/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Akinyemiju T, Abera S, Ahmed M, et al. The Burden of Primary Liver Cancer and Underlying Etiologies From 1990 to 2015 at the Global, Regional, and National Level: Results From the Global Burden of Disease Study 2015. JAMA Oncol 2017;3:1683-91. [Crossref] [PubMed]
- Villanueva A. Hepatocellular Carcinoma. N Engl J Med 2019;380:1450-62. [Crossref] [PubMed]
- Chen R, Xu X, Tao Y, et al. Exosomes in hepatocellular carcinoma: a new horizon. Cell Commun Signal 2019;17:1. [Crossref] [PubMed]
- Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378-90. [Crossref] [PubMed]
- Cheng AL, Kang YK, Chen Z, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol 2009;10:25-34. [Crossref] [PubMed]
- Kudo M, Finn RS, Qin S, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet 2018;391:1163-73. [Crossref] [PubMed]
- Lin J, Zhao H. Systemic management for patients with hepatobiliary tumors in a multi-dimensional view. Hepatobiliary Surg Nutr 2019;8:626-8. [Crossref] [PubMed]
- Miao R, Luo H, Zhou H, et al. Identification of prognostic biomarkers in hepatitis B virus-related hepatocellular carcinoma and stratification by integrative multi-omics analysis. J Hepatol 2014;61:840-9. [Crossref] [PubMed]
- Johnstone RM, Adam M, Hammond JR, et al. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem 1987;262:9412-20. [Crossref] [PubMed]
- Li L, Wang H. Heterogeneity of liver cancer and personalized therapy. Cancer Lett 2016;379:191-7. [Crossref] [PubMed]
- Roma-Rodrigues C, Raposo LR, Cabral R, et al. Tumor Microenvironment Modulation via Gold Nanoparticles Targeting Malicious Exosomes: Implications for Cancer Diagnostics and Therapy. Int J Mol Sci 2017;18:162. [Crossref] [PubMed]
- Chen G, Huang AC, Zhang W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018;560:382-6. [Crossref] [PubMed]
- Melo SA, Sugimoto H, O'Connell JT, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014;26:707-21. [Crossref] [PubMed]
- Li X, Li C, Zhang L, et al. The significance of exosomes in the development and treatment of hepatocellular carcinoma. Mol Cancer 2020;19:1. [Crossref] [PubMed]
- Miura K. The day will come to treat HCC in a drugstore? Hepatobiliary Surg Nutr 2017;6:420-3. [Crossref] [PubMed]
- Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med 1997;16:385-95. [Crossref] [PubMed]
- Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics 2000;56:337-44. [Crossref] [PubMed]
- Iasonos A, Schrag D, Raj GV, et al. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol 2008;26:1364-70. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Azizian NG, Li Y. XPO1-dependent nuclear export as a target for cancer therapy. J Hematol Oncol 2020;13:61. [Crossref] [PubMed]
- Zhang Y, Klein BJ, Cox KL, et al. Mechanism for autoinhibition and activation of the MORC3 ATPase. Proc Natl Acad Sci U S A 2019;116:6111-9. [Crossref] [PubMed]
- Fiorentino DF, Chung LS, Christopher-Stine L, et al. Most patients with cancer-associated dermatomyositis have antibodies to nuclear matrix protein NXP-2 or transcription intermediary factor 1γ. Arthritis Rheum 2013;65:2954-62. [Crossref] [PubMed]
- Liu T, Han X, Zheng S, et al. CALM1 promotes progression and dampens chemosensitivity to EGFR inhibitor in esophageal squamous cell carcinoma. Cancer Cell Int 2021;21:121. [Crossref] [PubMed]
- Paul D, Islam S, Manne RK, et al. F-box protein FBXO16 functions as a tumor suppressor by attenuating nuclear β-catenin function. J Pathol 2019;248:266-79. [Crossref] [PubMed]
- Zhu C, Chen X, Guan G, et al. IFI30 Is a Novel Immune-Related Target with Predicting Value of Prognosis and Treatment Response in Glioblastoma. Onco Targets Ther 2020;13:1129-43. [Crossref] [PubMed]
- Williams GH, Stoeber K. The cell cycle and cancer. J Pathol 2012;226:352-64. [Crossref] [PubMed]
- Ingham M, Schwartz GK. Cell-Cycle Therapeutics Come of Age. J Clin Oncol 2017;35:2949-59. [Crossref] [PubMed]
- Icard P, Fournel L, Wu Z, et al. Interconnection between Metabolism and Cell Cycle in Cancer. Trends Biochem Sci 2019;44:490-501. [Crossref] [PubMed]
- Ganapathy-Kanniappan S, Geschwind JF. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer 2013;12:152. [Crossref] [PubMed]
- Klutstein M, Nejman D, Greenfield R, et al. DNA Methylation in Cancer and Aging. Cancer Res 2016;76:3446-50. [Crossref] [PubMed]
- Morgan AE, Davies TJ, Mc Auley MT. The role of DNA methylation in ageing and cancer. Proc Nutr Soc 2018;77:412-22. [Crossref] [PubMed]
- Liu C, Chikina M, Deshpande R, et al. Treg Cells Promote the SREBP1-Dependent Metabolic Fitness of Tumor-Promoting Macrophages via Repression of CD8(+) T Cell-Derived Interferon-γ. Immunity 2019;51:381-97.e6. [Crossref] [PubMed]
- Marquardt JU, Galle PR, Teufel A. Molecular diagnosis and therapy of hepatocellular carcinoma (HCC): an emerging field for advanced technologies. J Hepatol 2012;56:267-75. [Crossref] [PubMed]
- Steuer CE, Ramalingam SS. Tumor Mutation Burden: Leading Immunotherapy to the Era of Precision Medicine? J Clin Oncol 2018;36:631-2. [Crossref] [PubMed]
- Zhang T, Merle P, Wang H, et al. Combination therapy for advanced hepatocellular carcinoma: do we see the light at the end of the tunnel? Hepatobiliary Surg Nutr 2021;10:180-92. [Crossref] [PubMed]
- Yang X, Xu H, Zuo B, et al. Downstaging and resection of hepatocellular carcinoma in patients with extrahepatic metastases after stereotactic therapy. Hepatobiliary Surg Nutr 2021;10:434-42. [Crossref] [PubMed]


