
Functional roles of epitranscriptomic marks in the cardiovascular system and disease: a narrative review
Introduction
Heart is the first organ to mature during the development. It sustains the live of organism by serving as an engine for blood circulation. The main cell types of the heart include cardiomyocytes (heart muscle), endothelial cells, fibroblasts, smooth muscle cells, pericytes, immune cells (myeloid and lymphoid), adipocytes, mesothelial cells, and neuronal cells (1). Unlike fish and amphibians, the mammalian cardiomyocytes do not normally regenerate themselves shortly after birth (2). Thus, the developmental studies of heart as well as how its dysfunction leads to cardiovascular disease are the topics of intensive studies. The protein-centered research could not uncover all the gene regulatory and signaling pathways to sustain the healthy heart. The emergence of non-protein-coding RNAs (ncRNAs), especially microRNAs (miRNAs) and more recent long non-coding RNAs (lncRNAs), has shed a light on further dissecting the gene regulatory networks of the heart (3,4). Yet, the deaths related to cardiovascular disease still ranks the top among all causes of deaths in industrialized countries (5), calling for further understanding of cardiac gene regulatory networks.
RNA is not only a mediator of genomic information encoded in DNA to the final products, proteins. The lifecycle of RNA is more dynamic, where they can be modified by a variety of enzymes. Indeed, more than 170 RNA modifications have been identified across species (6). The recent discoveries of RNA modifications and their importance in normal and pathophysiological conditions have led to the emergence of new field of study called, epitranscriptomics (7,8). This new field of study extends further understanding of epigenetics from the perspective of RNA, where the terminologies are adopted from epigenetics, including writers, readers, and erasers to categorize epitranscriptomic enzymes. These enzymes are active in many tissues and cell types, whose dyregulations are linked to a variety of disorders and critical diseases, such as cardiovascular disease (9,10). These epitranscriptomic enzymes mark all kinds of RNA species, including mRNAs and ncRNAs, such as ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), miRNAs, and lncRNAs (11). These marks are deposited by epitranscriptomic writers, interpreted by epitranscriptomic readers, and nullified by epitranscriptomic erasers. The lifecycle of RNA affected by these epitranscriptomic marks include splicing, subcellular localization, miRNA binding and biogenesis, mRNA stability and decay, and translation. Many of these marks are found throughout the transcriptome, which regulate downstream signaling pathways. Although modifications of rRNAs and tRNAs have been known over 50 years (12), only recently epitranscriptomic marks in mRNAs and ncRNAs have been identified. In this narrative review, we summarize the current status of epitranscriptomic marks in cardiovascular system and disease by focusing on three epitranscriptomic marks—N6-methyladenosine (m6A), adenosine to inosine (A-to-I) RNA editing, and 5-methylcytosine (m5C) as other epitranscriptomic marks are not studied extensively in cardiovascular system and disease. We present the following article in accordance with the Narrative Review reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1074/rc).
Methods
A literature review was performed using PubMed to search all scientific articles published through to January 1, 2022. The search terms used included: “cardiovascular”, “cardiovascular disease”, “epitranscriptomics”, “RNA modifications”, “2'-O-methylation”, “A-to-I RNA editing” “RNA editing”, “C-to-U RNA editing”, “ac4C”, “m1A”, “m3C”, “m5C”, “m6A”, “m7G”, and “pseudouridylation”. We use a table (Table 1) to present detailed search strategy.
Table 1
| Items | Specification |
|---|---|
| Date of search (specified to date, month and year) | January 2, 2022 |
| Databases and other sources searched | PubMed |
| Search terms used (including MeSH and free text search terms and filters) | “cardiovascular” “cardiovascular disease” “epitranscriptomics” “RNA modifications” “2'-O-methylation” “A-to-I RNA editing” “RNA editing” “C-to-U RNA editing” “ac4C” “m1A” “m3C” “m5C” “m6A” “m7G” “pseudouridylation” |
| Timeframe | July 1, 1980 – January 2, 2022 |
| Inclusion and exclusion criteria (study type, language restrictions etc.) | Inclusion: English, original articles |
| Exclusion: review articles | |
| Selection process (who conducted the selection, whether it was conducted independently, how consensus was obtained, etc.) | Shizuka Uchida |
| Any additional considerations, if applicable | Phenotypic and mechanistic studies |
N6-methyladenosine (m6A)
Among all RNA modifications, the most studied epitranscriptomic mark in recent years is N6-methyladenosine (m6A), which is the methylation of adenosine (A) at its nitrogen-6 position. This reversible modification is deposited by m6A writers [methyltransferase 3, N6-adenosine-methyltransferase complex catalytic subunit (METTL3), methyltransferase 14, N6-adenosine-methyltransferase complex catalytic subunit (METTL4), WT1 associated protein (WTAP), vir like m6A methyltransferase associated (VIRMA, also known as KIAA1429), methyltransferase 16, N6-methyladenosine (METTL16), RNA binding motif protein 15 (RBM15), RNA binding motif protein 15B (RBM15B), and zinc finger CCCH-type containing 13 (ZC3H)], interpreted by m6A readers [YTH N6-methyladenosine RNA binding proteins 1 (YTHDF1), YTH N6-methyladenosine RNA binding proteins 2 (YTHDF2), YTH N6-methyladenosine RNA binding proteins 3 (YTHDF3), YTH domain containing 1 (YTHDC1), YTH domain containing 2 (YTHDC2), heterogeneous nuclear ribonucleoprotein C (HNRNPC), RNA binding motif protein X-linked (RBMX, also known as HNRNPG), insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1), insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2), and insulin like growth factor 2 mRNA binding protein 3 (IGF2BP3)], and nullified by m6A erasers [alkB homolog 5, RNA demethylase (ALKBH5) and FTO alpha-ketoglutarate dependent dioxygenase (FTO)] (13,14) (Figure 1). The m6A sites are found mostly in mRNAs, especially around stop codon (15), and regulate mRNA metabolism, such as splicing, nuclear export, mRNA stability, and translation (16). Because of the availability of high-throughput methods to detect m6A sites {e.g., m6A-seq [also more commonly known as m6A RNA immunoprecipitation followed by high-throughput sequencing (MeRIP-seq)], m6A individual-nucleotide-resolution cross-linking and immunoprecipitation (miCLIP), and m6A crosslinking immunoprecipitation sequencing (m6A-CLIP); comprehensively reviewed and compared in (17)}, a number of m6A profiles were reported in healthy hearts (18) as well as in those compared to diseased ones and/or cardiovascular disease models in mice and rats, including during cardiac development (19,20), cardiomyopathy (21), diabetic cardiomyopathy (22), heart failure (21,23), intermittent fasting in high-fat diet-induced cardiomyopathy (24), and myocardial infarction (25). Although these screening data are informative for the global changes in m6A profiles under different cardiac conditions, gain/loss-of-function analysis of each m6A enzyme focusing on cardiac physiology and disease is important to understand the function of each m6A enzyme, which are described below (Table 2).
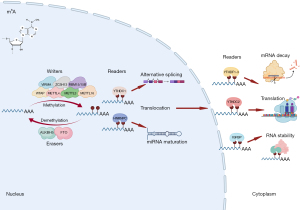
Table 2
| Epitrancriptomic enzyme | Experimental system | Phenotypes/mechanisms | Reference |
|---|---|---|---|
| m6A writer, Mettl3 | Neonatal rat ventricular cardiomyocytes, cardiomyocyte-specific Mettl3 conditional knockout mice | Causes cardiac hypertrophy | (26) |
| m6A reader, Ythdc1 | Cardiomyocyte-specific Ythdc1 conditional knockout mice | Possibly required for the proper splicing of sarcomeric protein, Titin | (27) |
| m6A reader, Ythdf2 | Ythdf2 overexpressing mice | Suppresses cardiac hypertrophy by recognizing m6A site on the Myh7 mRNA | (28) |
| m6A eraser, Fto | Fto overexpressing mice | Demethylates cardiac contractile mRNAs to prevent their mRNA degradation and promote their protein expression to preserve cardiac functions in the infarcted hearts | (29) |
| m6A eraser, Alkbh5 | Alkbh5 knockout and overexpressing mice, hiPSC-CM | Stabilizes the m6A reader YTHDF1 mRNA in a m6A dependent manner, thereby promoting the translation of YAP | (30) |
| A-to-I RNA editing writer, Adar1 | Adar1 knockout mice | Results embryonic death due to massive apoptosis and aberrant interferon induction | (31) |
| A-to-I RNA editing writer, Adar1 | Cardiac-specific Adar1 conditional knockout mice | Regulates the cardiomyocyte survival and proliferation | (32) |
| A-to-I RNA editing writer, Adar1 | Cardiomyocyte-specific Adar1 conditional knockout mice | Results in increased lethality due to increased endoplasmic stress leading to apoptosis and reduction in miRNA levels | (33) |
| A-to-I RNA editing writer, Adar2 | Neonatal rat cardiomyocytes, cardiomyocyte-specific Adar2 overexpresing mice | Negatively regulates mature miR-34a to protect murine hearts from acute myocardial infarction | (34) |
| m5C writer, Trdmt1 (Dnmt2) | Dnmt2 mutant mice | Results in cardiac hypertrophy possibly due to decreased methylation and increased dissociation of small nuclear RNA from P-TEFb complex | (35) |
| m5C writer, Nsun4 | Muscle-specific Nsun4 conditional knockout mice | Methylates 12S rRNA and forms a complex with MTERF4 to regulated mitoribosomal assembly | (36) |
| m5C writer, Nsun2 | TALEN-mediated Nsun2 knockout rats | Methylates Icam1 mRNA to promote its translation | (37) |
m6A writers
Pathological cardiac hypertrophy is a condition in which the heart enlarges in response to stresses, such as pressure overload or myocardial infarction, which progresses to heart failure (38). Understanding the cascade of events in maladaptive hearts is important for therapeutic purpose. Interestingly, the m6A level increased in hypertrophied cardiomyocytes (26). When a major component of the m6A methyltransferase complex, Mettl3, was overexpressed in neonatal rat ventricular cardiomyocytes, cardiomyocytes were hypertrophied, while the opposite effect was observed when silencing Mettl3. When Mettl3 was overexpressed in mice, cardiac hypertrophy was observed, while Mettl3 floxed mice crossed with the cardiomyocyte-specific cre-driver, Myh7-cre (also known as β-MHC cre), displayed signs of heart failure with aging and stress, suggesting the functional importance of Mettl3 in cardiac physiology.
m6A readers
Just as other readers of epitranscriptomic marks, m6A readers are RNA-binding proteins (RBP), which affects the mRNA stability and decay, leading to translational regulations (39). As these readers bind many RNAs, it is often difficult to pinpoint only one transcript been affected in a particular condition. Nevertheless, there are several targeted studies of m6A readers in cardiovascular system and disease. For example, when Ythdc1 floxed mice were crossed with the constitutive active, cardiomyocyte-specific cre-driver, α-MHC-Cre, homozygous Ythdc1 conditional knockout mice died around 10 weeks of age (27), suggesting its importance in postnatal cardiac function. The detailed analysis showed that Ythdc1-ablated mice displayed left ventricular chamber enlargement and severe systolic dysfunction, which are the hallmark of dilated cardiomyopathy, as well as decrease of cardiomyocyte contractility and disordered sarcomere arrangement. Mechanistically, it was found that Ythdc1 might be required for the proper splicing of sarcomeric protein, Titin.
Another m6A reader, YTHDF2, is also indicated to be involved in cardiac physiology. In both human and mice, the expression of YTHDF2 mRNA and protein, but not its family members—YTHDF1 or YTHDF3, are upregulated during the progression of heart failure (28). When challenged with transverse aortic constriction, Ythdf2 overexpressing mice attenuated cardiac hypertrophy. Mechanistically, YTHDF2 suppresses cardiac hypertrophy by recognizing m6A site on the myosin heavy chain 7 (Myh7) mRNA (also known as β-MHC, which is a marker for cardiac hypertrophy) to promote its degradation.
m6A erasers
Discovered as human obesity-susceptibility gene (40,41), FTO (fat mass and obesity associated gene) is a m6A demethylase that is shown to be involved in cardiac physiology. In failing human, pig, and murine hearts, the expression of FTO was decreased, which coincided with the increased level of m6A marks and decreased cardiomyocyte contractile function (29). When challenged with a permanent ligation of the left anterior descending artery (a mouse model of myocardial infarction), the hearts of Fto overexpressing mice attenuated the pathological features of failing hearts (e.g., decreased cardiac contractile function). Mechanistically, Fto demethylates cardiac contractile mRNAs [e.g., natriuretic peptide A (Nppa, also known as ANP), Myh7, ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 (Atp2a2, also known as Serca2a), and ryanodine receptor 2, cardiac (Ryr2)] to prevent their mRNA degradation and promote their protein expression to preserve cardiac functions in the infarcted hearts. However, there is still an ongoing debate as to the validity of FTO as m6A eraser as it targets N6,2'-O-dimethyladenosine (m6Am, which is a modification found adjacent to the mRNA cap) instead of m6A (42). Thus, more research is needed to firmly confirm the functional role of FTO in the context of m6A marks.
Although the adult mammalian cardiomyocytes are terminally differentiated without capacity for regeneration, neonatal murine hearts can regenerate within the first week after birth when challenged with apical resection or myocardial infarction, mediated by cardiomyocyte proliferation (43). A recent study shows that the m6A eraser, ALKBH5, plays a role in cardiac regeneration as the neonatal Alkbh5 knockout mice fail to regenerate their hears after apical resection, while overexpression of Alkbh5 in juvenile (7 days old) and adult (8 weeks old) mice promoted cardiomyocyte proliferation after myocardial infarction (30). In vitro, overexpression of ALKBH5 enhanced mitosis in human induced pluripotent stem cell-derived cardiomyocytes. Mechanistically, ALKBH5 stabilizes the m6A reader YTHDF1 mRNA in a m6A dependent manner, thereby promoting the translation of Yes1 associated transcriptional regulator (YAP1, also known as YAP), which is a downstream effector of the Hippo pathway that is important for cardiomyocyte growth.
A-to-I RNA editing
Adenosine to inosine (A-to-I) RNA editing is a conversion of A into I via the catalytic reaction of adenosine deaminase acting on RNAs (ADARs) by binding to double-stranded RNA (44) (Figure 2). RNA editing affects RNA metabolism by alterning splicing, miRNA biogenesis and binding, amino acid conversion, lncRNA binding and structures, RNA stability, and translation. Compared to m6A marks, RNA editing occurs most frequently in introns and 3’-untranslated regions (UTR) as both epitranscriptomic marks target the same nitrogen-6 position of A (45); thus, they are in a reciprocal relation. Unlike m6A, RNA editing is an irreversible process with no known reader to date. In mammals, there are three ADAR genes: ADAR (ADAR1), ADARB1 (ADAR2), and catalytically inactive ADARB2 (ADAR3). The mutation in the human ADAR1 gene is known to cause of the Aicardi-Goutières autoimmune disease (46), while the accumulation of ADAR1 protein was observed in atherosclerotic plaques (47), which are the most common cause of coronary artery diseases. In mice, the whole body knockout of Adar1 results in embryonic death due to massive apoptosis and aberrant interferon induction (31), which can be rescued to live birth by ablating the RIG-I-like receptors, mitochondrial antiviral signaling protein (Mavs) or interferon induced with helicase C domain 1 (Ifih1, also known as Mda5) (48-50). Interestingly, these massive apoptotic events are most visible in the embryonic heart, which the recent study crossing Adar1 conditional knockout mice with the early cardiac specific cre-driver, Nkx2.5-Cre, resulted in embryonic death due to increased apoptosis and reduced proliferation in developing cardiomyocytes (32) (Table 2), suggesting that Adar1 is important in regulating the cardiomyocyte survival and proliferation. Because NK2 homeobox 5 (Nkx2-5) gene is also expressed in other cell type (i.e., endothelium) and tissue types (tongue, thymus, and spleen) during embryonic development (51), it is possible that the ablation of Adar1 in early cardiac field may not be specific for cardiomyocytes. However, another recent study crossing Adar1 conditional knockout mice with the endothelial-specific cre driver, VE-Cadherin-Cre (Chd5-Cre), demonstrated that the ablating Adar1 in endothelial cells did not result in embryonic death, instead postnatal death due to growth retardation within three weeks after birth for 75% of neonates (52). To further confirm the important role of Adar1 in cardiomyocytes, another study crossing Adar1 conditional knockout mice with the tamoxifen-inducible cardiomyocyte specific cre-driver, αMHC-MCM, showed that ablating Adar1 in adult cardiomyocytes resulted in increased lethality due to increased endoplasmic stress leading to apoptosis and reduction in miRNA levels, especially miR-199a-5p that regulates unfolded protein response (UPR) (33). Taken together, these studies demonstrate that Adar1 in murine cardiomyocytes is important for development and physiology of the heart.
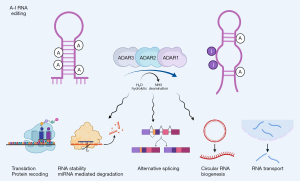
Compared to ADAR1, ADAR2 is more specific in its targets [i.e., glutamate ionotropic receptor AMPA type subunit 2 (GRIA2, also known as GluR2) pre-mRNA editing at Q/R site (53)], which demonstrated to be very useful for RNA-based genome editing by fusing the catalytic domain of ADAR2 to CRISPR-Cas13b enzyme to perform A-to-I conversion in RNA transcripts (54). In mice, the expression of Adar2 was induced in exercised hearts (34). In vitro, overexpression of Adar2 inhibited doxorubicin-induced apoptosis in neonatal rat cardiomyocytes, while silencing of Adar2 resulted in increased apoptosis. When Adar2 was overexpressed in a cardiac specific manner, it protected murine hearts from acute myocardial infarction by negatively regulating mature miR-34a, suggesting that Adar2 could be a potential therapeutic target for cardiovascular disease (34).
5-methylcytosine (m5C)
Just as DNA, RNA can be marked by 5-methylcytosine (m5C), which are found abundantly in rRNAs and tRNAs but also in mRNAs and ncRNAs, which stabilize RNA folding and structures (55,56). This reversible modification is deposited by m5C writers [NOP2 nucleolar protein (NOP2, also known as (NSUN1), NOP2/Sun RNA methyltransferase 2 (NSUN2), NOP2/Sun RNA methyltransferase 3 (NSUN3), NOP2/Sun RNA methyltransferase 4 (NSUN4), NOP2/Sun RNA methyltransferase 5 (NSUN5), NOP2/Sun RNA methyltransferase 6 (NSUN6), NOP2/Sun RNA methyltransferase family member 7 (NSUN7), and tRNA aspartic acid methyltransferase 1 (TRDMT1, also known as DNMT2)], interpreted by m5C readers [Aly/REF export factor (ALYREF), Y-box binding protein 1 (YBX1), and RAD52 homolog, DNA repair protein (RAD52)], and nullified by m5C erasers [tet methylcytosine dioxygenase 1 (TET1), tet methylcytosine dioxygenase 2 (TET2), and tet methylcytosine dioxygenase 3 (TET3)] (Figure 3). By employing RNA bisulfite sequencing (RNA-BisSeq), 3,575 m5C sites in 1,574 mRNAs were identified in the murine heart (57), although differential m5C analysis of diseased hearts compared to the healthy ones has not been performed yet.
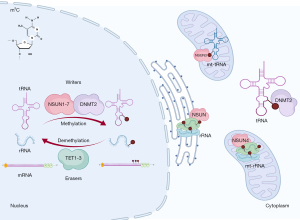
The DNA methyltransferase homologue, TRDMT1 (DNMT2), methylates tRNAs rather than DNA (58). Although Dnmt2 mutated flies, plants, and mice are viable and fertile, Dnmt2 morphant zebrafish embryos showed developmental defects in the brain, retina, and liver (59). When the adult (3 months old) Dnmt2 mutant mice were examined carefully, cardiac hypertrophy due to induced cardiac growth with preserved function was observed (35) (Table 2). This phenotype was possibly due to decreased methylation and increased dissociation of small nuclear RNA, RNA, 7SK, nuclear (Rn7sk), from P-TEFb complex (positive transcription elongation factor), thereby over-activating RNA polymerase II to induce cardiac hypertrophy via gene expression.
Another m5C writer, Nsun4, is also important in cardiomyocytes. Crossing Nsun4 conditional knockout mice with the muscle-specific (both cardiac and skeletal muscles) cre-driver, Ckmm-Cre, it was found that ablation of Nsun4 resulted in shorter lifespan (death within the age of 25 weeks) with progressive cardiomyopathy based on the increased heart to body weight ratio, leading to mitochondrial dysfunction caused by impaired biogenesis of the respiratory chain complexes (36). Mechanistically, Nsun4 is a dual function protein, which methylates 12S rRNA and forms a complex with the mitochondrial protein, mitochondrial transcription termination factor 4 (MTERF4), to regulated mitoribosomal assembly.
Besides cardiomyocytes, m5C marks are also important in endothelial functions. Using TALEN-mediated knockout of m5C writer, Nsun2, in rats demonstrated that ablation of Nsun2 impaired the formation of allograft arteriosclerosis (hardening of the arteries) in a model of aortic allograft by transplanting thoracic aortic allografts of wildtype or Nsun2 knockout rats into the abdominal aorta of donor rats, suggesting that Nsun2 is important for neointima formation (37). Mechanistically, Nsun2 methylates intercellular adhesion molecule 1 (Icam1) mRNA to promote its translation, thereby increasing the adhesion of leukocytes to endothelial cells to regulate vascular endothelial inflammation and atherosclerosis.
Conclusions
Compared to m6A, A-to-I RNA editing, and m5C, less is known about other epitranscriptomic marks in the heart. Because of the convenience and availability of next generation sequencing, especially RNA sequencing (RNA-seq), high-throughput screening studies should be carried out to understand the changes in epitranscriptomic marks in diseased hearts compared to the healthy ones, for example. Furthermore, more genetic studies, especially those using cell-type-specific knockout mice (60-62), are urgently needed to understand functional roles of epitranscriptomic marks in the heart. Because of the availability of induced pluripotent stem cells (iPSC) and their differentiated cell types (i.e., iPSC-derived cardiomyocytes), more functional studies of epitranscriptomic enzymes should be performed by gain/loss-of-function experiments, especially using CRISPR/Cas9 system to ablate each epitranscriptomic enzyme in iPSC. These functional and mechanistic studies along with screening studies are needed to better characterize epitranscriptomic marks in cardiovascular system and disease.
Acknowledgments
Funding: This work was supported by a grant from the Novo Nordisk Foundation (No. NNF18OC0033438).
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1074/rc
Peer Review File: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1074/prf
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1074/coif). SU serves as an unpaid editorial board member of Annals of Translational Medicine from November 2021 to October 2023. MI has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Litviňuková M, Talavera-López C, Maatz H, et al. Cells of the adult human heart. Nature 2020;588:466-72. [Crossref] [PubMed]
- Lázár E, Sadek HA, Bergmann O. Cardiomyocyte renewal in the human heart: insights from the fall-out. Eur Heart J 2017;38:2333-42. [Crossref] [PubMed]
- Uchida S, Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ Res 2015;116:737-50. [Crossref] [PubMed]
- Thum T, Galuppo P, Wolf C, et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation 2007;116:258-67. [Crossref] [PubMed]
- WHO. Cardiovascular diseases (CVDs). 2021. Available online: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds)
- Boccaletto P, Machnicka MA, Purta E, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 2018;46:D303-7. [Crossref] [PubMed]
- Roundtree IA, He C. RNA epigenetics--chemical messages for posttranscriptional gene regulation. Curr Opin Chem Biol 2016;30:46-51. [Crossref] [PubMed]
- Saletore Y, Meyer K, Korlach J, et al. The birth of the Epitranscriptome: deciphering the function of RNA modifications. Genome Biol 2012;13:175. [Crossref] [PubMed]
- Sikorski V, Karjalainen P, Blokhina D, et al. Epitranscriptomics of Ischemic Heart Disease-The IHD-EPITRAN Study Design and Objectives. Int J Mol Sci 2021;22:6630. [Crossref] [PubMed]
- Uchida S, Jones SP. RNA Editing: Unexplored Opportunities in the Cardiovascular System. Circ Res 2018;122:399-401. [Crossref] [PubMed]
- Esteller M, Pandolfi PP. The Epitranscriptome of Noncoding RNAs in Cancer. Cancer Discov 2017;7:359-68. [Crossref] [PubMed]
- Zamecnik PC. Summary of symposium on transfer RNA and transfer RNA modification in differentiation and neoplasia. Cancer Res 1971;31:716-21. [PubMed]
- Fu Y, Dominissini D, Rechavi G, et al. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet 2014;15:293-306. [Crossref] [PubMed]
- Yang Y, Hsu PJ, Chen YS, et al. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res 2018;28:616-24. [Crossref] [PubMed]
- Jiang X, Liu B, Nie Z, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther 2021;6:74. [Crossref] [PubMed]
- Wang S, Lv W, Li T, et al. Dynamic regulation and functions of mRNA m6A modification. Cancer Cell Int 2022;22:48. [Crossref] [PubMed]
- Capitanchik C, Toolan-Kerr P, Luscombe NM, et al. How Do You Identify m6 A Methylation in Transcriptomes at High Resolution? A Comparison of Recent Datasets. Front Genet 2020;11:398. [Crossref] [PubMed]
- Xiong X, Hou L, Park YP, et al. Genetic drivers of m6A methylation in human brain, lung, heart and muscle. Nat Genet 2021;53:1156-65. [Crossref] [PubMed]
- Yang Y, Shen S, Cai Y, et al. Dynamic Patterns of N6-Methyladenosine Profiles of Messenger RNA Correlated with the Cardiomyocyte Regenerability during the Early Heart Development in Mice. Oxid Med Cell Longev 2021;2021:5537804. [Crossref] [PubMed]
- Yang C, Zhao K, Zhang J, et al. Comprehensive Analysis of the Transcriptome-Wide m6A Methylome of Heart via MeRIP After Birth: Day 0 vs. Day 7. Front Cardiovasc Med 2021;8:633631. [Crossref] [PubMed]
- Berulava T, Buchholz E, Elerdashvili V, et al. Changes in m6A RNA methylation contribute to heart failure progression by modulating translation. Eur J Heart Fail 2020;22:54-66. [Crossref] [PubMed]
- Ju W, Liu K, Ouyang S, et al. Changes in N6-Methyladenosine Modification Modulate Diabetic Cardiomyopathy by Reducing Myocardial Fibrosis and Myocyte Hypertrophy. Front Cell Dev Biol 2021;9:702579. [Crossref] [PubMed]
- Hinger SA, Wei J, Dorn LE, et al. Remodeling of the m6A landscape in the heart reveals few conserved post-transcriptional events underlying cardiomyocyte hypertrophy. J Mol Cell Cardiol 2021;151:46-55. [Crossref] [PubMed]
- Xu Z, Qin Y, Lv B, et al. Intermittent Fasting Improves High-Fat Diet-Induced Obesity Cardiomyopathy via Alleviating Lipid Deposition and Apoptosis and Decreasing m6A Methylation in the Heart. Nutrients 2022;14:251. [Crossref] [PubMed]
- Su X, Shen Y, Jin Y, et al. Aging-Associated Differences in Epitranscriptomic m6A Regulation in Response to Acute Cardiac Ischemia/Reperfusion Injury in Female Mice. Front Pharmacol 2021;12:654316. [Crossref] [PubMed]
- Dorn LE, Lasman L, Chen J, et al. The N6-Methyladenosine mRNA Methylase METTL3 Controls Cardiac Homeostasis and Hypertrophy. Circulation 2019;139:533-45. [Crossref] [PubMed]
- Gao S, Sun H, Chen K, et al. Depletion of m6 A reader protein YTHDC1 induces dilated cardiomyopathy by abnormal splicing of Titin. J Cell Mol Med 2021;25:10879-91. [Crossref] [PubMed]
- Xu H, Wang Z, Chen M, et al. YTHDF2 alleviates cardiac hypertrophy via regulating Myh7 mRNA decoy. Cell Biosci 2021;11:132. [Crossref] [PubMed]
- Mathiyalagan P, Adamiak M, Mayourian J, et al. FTO-Dependent N6-Methyladenosine Regulates Cardiac Function During Remodeling and Repair. Circulation 2019;139:518-32. [Crossref] [PubMed]
- Han Z, Wang X, Xu Z, et al. ALKBH5 regulates cardiomyocyte proliferation and heart regeneration by demethylating the mRNA of YTHDF1. Theranostics 2021;11:3000-16. [Crossref] [PubMed]
- Wang Q, Miyakoda M, Yang W, et al. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem 2004;279:4952-61. [Crossref] [PubMed]
- Moore JB 4th. The A-to-I RNA Editing Enzyme Adar1 Is Essential for Normal Embryonic Cardiac Growth and Development. Circ Res 2020;127:550-2. [Crossref] [PubMed]
- El Azzouzi H, Vilaça AP, Feyen DAM, et al. Cardiomyocyte Specific Deletion of ADAR1 Causes Severe Cardiac Dysfunction and Increased Lethality. Front Cardiovasc Med 2020;7:30. [Crossref] [PubMed]
- Wu X, Wang L, Wang K, et al. ADAR2 increases in exercised heart and protects against myocardial infarction and doxorubicin-induced cardiotoxicity. Mol Ther 2022;30:400-14. [Crossref] [PubMed]
- Ghanbarian H, Wagner N, Polo B, et al. Dnmt2/Trdmt1 as Mediator of RNA Polymerase II Transcriptional Activity in Cardiac Growth. PLoS One 2016;11:e0156953. [Crossref] [PubMed]
- Metodiev MD, Spåhr H, Loguercio Polosa P, et al. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet 2014;10:e1004110. [Crossref] [PubMed]
- Luo Y, Feng J, Xu Q, et al. NSun2 Deficiency Protects Endothelium From Inflammation via mRNA Methylation of ICAM-1. Circ Res 2016;118:944-56. [Crossref] [PubMed]
- Shimizu I, Minamino T. Physiological and pathological cardiac hypertrophy. J Mol Cell Cardiol 2016;97:245-62. [Crossref] [PubMed]
- Grosset C, Chen CY, Xu N, et al. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 2000;103:29-40. [Crossref] [PubMed]
- Dina C, Meyre D, Gallina S, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet 2007;39:724-6. [Crossref] [PubMed]
- Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007;316:889-94. [Crossref] [PubMed]
- Mauer J, Jaffrey FTO SR. m6 Am, and the hypothesis of reversible epitranscriptomic mRNA modifications. FEBS Lett 2018;592:2012-22. [Crossref] [PubMed]
- Lam NT, Sadek HA. Neonatal Heart Regeneration: Comprehensive Literature Review. Circulation 2018;138:412-23. [Crossref] [PubMed]
- Vesely C, Jantsch MF. An I for an A: Dynamic Regulation of Adenosine Deamination-Mediated RNA Editing. Genes (Basel) 2021;12:1026. [Crossref] [PubMed]
- Xiang JF, Yang Q, Liu CX, et al. N6-Methyladenosines Modulate A-to-I RNA Editing. Mol Cell 2018;69:126-135.e6. [Crossref] [PubMed]
- Rice GI, Kasher PR, Forte GM, et al. Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat Genet 2012;44:1243-8. [Crossref] [PubMed]
- Stellos K, Gatsiou A, Stamatelopoulos K, et al. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat Med 2016;22:1140-50. [Crossref] [PubMed]
- Pestal K, Funk CC, Snyder JM, et al. Isoforms of RNA-Editing Enzyme ADAR1 Independently Control Nucleic Acid Sensor MDA5-Driven Autoimmunity and Multi-organ Development. Immunity 2015;43:933-44. [Crossref] [PubMed]
- Liddicoat BJ, Piskol R, Chalk AM, et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 2015;349:1115-20. [Crossref] [PubMed]
- Mannion NM, Greenwood SM, Young R, et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep 2014;9:1482-94. [Crossref] [PubMed]
- Moses KA, DeMayo F, Braun RM, et al. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis 2001;31:176-80. [Crossref] [PubMed]
- Guo X, Liu S, Yan R, et al. ADAR1 RNA editing regulates endothelial cell functions via the MDA-5 RNA sensing signaling pathway. Life Sci Alliance 2022;5:e202101191. [Crossref] [PubMed]
- Higuchi M, Single FN, Köhler M, et al. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell 1993;75:1361-70. [Crossref] [PubMed]
- Cox DBT, Gootenberg JS, Abudayyeh OO, et al. RNA editing with CRISPR-Cas13. Science 2017;358:1019-27. [Crossref] [PubMed]
- Xue C, Zhao Y, Li L. Advances in RNA cytosine-5 methylation: detection, regulatory mechanisms, biological functions and links to cancer. Biomark Res 2020;8:43. [Crossref] [PubMed]
- Trixl L, Lusser A. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip Rev RNA 2019;10:e1510. [Crossref] [PubMed]
- Yang X, Yang Y, Sun BF, et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res 2017;27:606-25. [Crossref] [PubMed]
- Ehrenhofer-Murray AE. Cross-Talk between Dnmt2-Dependent tRNA Methylation and Queuosine Modification. Biomolecules 2017;7:14. [Crossref] [PubMed]
- Tuorto F, Herbst F, Alerasool N, et al. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J 2015;34:2350-62. [Crossref] [PubMed]
- Plikus MV, Wang X, Sinha S, et al. Fibroblasts: Origins, definitions, and functions in health and disease. Cell 2021;184:3852-72. [Crossref] [PubMed]
- Payne S, De Val S, Neal A. Endothelial-Specific Cre Mouse Models. Arterioscler Thromb Vasc Biol 2018;38:2550-61. [Crossref] [PubMed]
- Doetschman T, Azhar M. Cardiac-specific inducible and conditional gene targeting in mice. Circ Res 2012;110:1498-512. [Crossref] [PubMed]


