
Next-generation sequencing and targeted quantitative real-time polymerase chain reaction for detection of Akebiae Caulis in the traditional Chinese medical formula Longdan Xiegan Wan
Introduction
Traditional Chinese Medicine (TCM) was included in the 11th revision of the World Health Organization (WHO) International Statistical Classification of Diseases and Related Health Problems (ICD) which came into effect on 1 January 2022 for the first time. This inclusion suggests that TCM will become more affordable and readily available than Western medicine in some countries.
Traditional medicine, dietary supplements, and natural herbal products containing herbal blends have flooded the markets in many Western countries (1). The public relies on drug labels for drug information (2). Regulators require product labels to include all product information. However, Chinese herbal medicinal products may be adulterated with impurities or contaminants during cultivation and processing (3). The general methods which can detect specific compounds in TCM are recorded in the Pharmacopoeia of the People’s Republic of China (2015 Edition) (4), and are based on morphology, microscopic identification, thin-layer chromatography (TLC), and high-performance liquid chromatography (HPLC). However, these methods cannot detect all labeled and unlabeled species simultaneously in multi-ingredient TCMs (4,5). A study showed that only 68% of Hypericum perforatum herbal products contained the target species, and differences between ingredient species and those listed on the label were detected in all products (6). To improve the safety and effectiveness of TCM, regulatory agencies should seek more effective and comprehensive detection methods.
Next-generation sequencing (NGS) based on DNA barcoding has been successfully used to identify components in mixed samples of animals and plants (7-9). There have also been cases of TCM component identification based on quantitative real-time polymerase chain reaction (qPCR) (8). However, the DNA of species in TCM compete with DNA barcode primers in the PCR process, where the more common templates are more easily amplified. This results in false negatives for low-abundance species (10,11). However, qPCR primers only specifically bind to the target gene, providing the possibility of detecting low-abundance species. To avoid the possible false-negative results of NGS, we also applied qPCR to identify the Akebiae Caulis (Mu Tong) in Longdan Xiegan Wan (LDXGW), which provided a method for the detection of Akebiae Caulis in the compound.
In the present study, we attempted to detect TCM that contain Akebiae Caulis. Aristolochiae Manshuriensis Caulis (Guan Mutong) and Clematidis Armandii Caulis (Chuan Mutong), which are nephrotoxic and carcinogenic, are often misused as Akebiae Caulis (12,13). However, the 2015 edition of the Chinese Pharmacopoeia did not describe a detection method of Akebiae Caulis in the TCM compounds (3). The misuse of the Akebiae Caulis, Clematidis Armandii Caulis, and Aristolochiae Manshuriensis Caulis will cause serious safety issues (14-16).
We performed DNA barcoding using NGS and targeted qPCR for Akebiae Caulis detection. Three primer pairs of qPCR which are specific for Akebiae Caulis, Clematidis Armandii Caulis, and Aristolochiae Manshuriensis Caulis were used in our analysis. A single primer pair of internal transcribed spacer 2 (ITS2) was employed for DNA barcoding, as ITS2 is a standard molecular marker with a high sensitivity for species identification (17-20). We selected LDXGW randomly, which should include Akebiae Caulis, as the research object. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-2415/rc).
Methods
Sample collection
Six dry stem specimens of the botanical species source of Clematidis Armandii caulis (Chuan Mutong), Akebiae Caulis (Mu Tong), and Aristolochiae Manshuriensis Caulis (Guan Mutong) were collected from the Yunnan, Guangxi, Chongqing, Hubei, and Liaoning provinces to obtain the reference ITS2 sequences to design qPCR primers (Table 1). Commercial LDXGW specimens were randomly purchased from Beijing Tong Ren Tang Drugstore (Lot No. 17082007; Luohe, Henan, China), and the commercial product was marked as LDXGW139. The other LDXGW samples were purchased from Tianyi Qinkun Pharmaceutical Co., Ltd. (Lot No. 180801; Xi’an, China) and Lanzhou Foci Pharmaceutical Co., Ltd. (Lot No. 18I33; Lanzhou, China) were marked as LDXGW138 and LDXGW137, respectively.
Table 1
| Herbal material | Species | Location | ITS2 |
|---|---|---|---|
| Chuan Mutong | Clematis armandii | Yunnan | MK558851 |
| Clematis montana | Guangxi | MK558852 | |
| Mu Tong | Akebia quinata | Chongqing | MK558847 |
| Akebia trifoliata | Hubei | MK558848 | |
| Akebia trifoliata var. australis | Hubei | MK558849 | |
| Guan Mutong | Aristolochia manshuriensis | Liaoning | MK558850 |
ITS2, internal transcribed spacer 2.
NGS and qPCR analysis
The methods for NGS and qPCR analysis were described in previous study (6). In brief, DNA extraction, amplification, and library construction were performed using commercial kits. Ion S5 sequencing platform and Ion 530 chip were used to generate high-quality (HQ) reads of targeted amplicons. We performed qPCR using the ViiA7 Real-Time System (Applied Biosystems®), whereby a standard qPCR protocol was employed. Details are provided in the supplementary methods.
Results
Specimen authentication, ITS2 sequence variation, and qPCR primer design
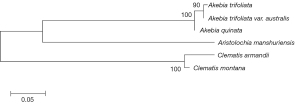
The ITS2 regions of specimens were all successfully amplified and bi-directionally sequenced using Sanger sequencing technology. Specimen authentications were implemented in the DNA barcoding system of Chinese herbal materials (21). The results confirmed that the samples were of the species recorded in the Chinese Pharmacopoeia 2015 edition (Chuan Mutong, Mu Tong) and 1990 edition (Guan Mutong). The sequence lengths of Chuan Mutong, Mu Tong, and Guan Mutong were 218, 216, and 274 bp, respectively. There are 123 variable position sites and 76 Parsim-info position sites among Chuan Mutong, Mu Tong, and Guan Mutong, so there are plenty of options to design qPCR primers to detect these three herbal materials. In addition, the phylogenetic tree strongly supported the potential discrimination of Chuan Mutong, Mu Tong, and Guan Mutong using ITS2 regions (Figure 1).
Targeted qPCR detections
Targeted qPCR detected Akebiae Caulis (Mu Tong) in three LDXGW samples, and one brand of LDXGW was not detected for Akebiae Caulis. Clematidis Armandii Caulis (Chuan Mutong) was detected in all commercial samples, while Aristolochiae Manshuriensis Caulis (Guan Mutong) was negative in all commercial samples (Figure 2). The information of the qPCR primer pairs is summarized in Table 2.

Table 2
| No. | Species | Forward primer (5' to 3') | Reverse primer (5' to 3') |
|---|---|---|---|
| 1 | Akebiae Caulis | TGGTGGTTGACGTGCCTCTT | CACCGACAAGGTCCGCGT |
| 2 | Clematidis Armandii Caulis | GCGGTCAGCGGTGGTTGT | CGCGTCGTTTCTGTCTTTGG |
| 3 | Aristolochiae Manshuriensis Caulis | TCGGGTGCGGTTGGCT | GGAGGCGAACGGTTAGGGTC |
qPCR, quantitative real-time polymerase chain reaction.
NGS results of complex matrix
Based on the Ion Torrent S5 sequencing platform, three batches of LDXGW samples (LDXGW137, LDXGW138, and LDXGW139) were used for NGS of ITS2 molecular tags in this study, and a total of 3,035,227 reads were obtained. A total of 1,957,902 HQ reads were obtained after filtering adaptors, low-quality sequences [quality value (QV) < quality score of 20 (Q20)], and short reads (length <300 bp). The filtered data were subjected to TCM V1.0 plug of Ion Torrent S5-in analysis, and Clematis peterae (Tie-Xian Lian) was detected in all commercial samples (LDXGW137, LDXGW138, LDXGW139) based on the alignment of ITS2 sequences. In contrast, Akebiae Caulis was not found in any of the three samples. The non-prescribed species, Clematis armandii, and Clematis montana, were detected in LDXGW139.
NGS versus qPCR for Akebiae Caulis and adulterants detection
Although there are 15 kinds of TCM compounds containing Akebiae Caulis in the 2015 edition of the Chinese Pharmacopoeia, an identification method of Akebiae Caulis is not specified (3). Aristolochic acid (AA), which is found in many Aristolochiaceae species, is nephrotoxic (20). Aristolochiae Manshuriensis Caulis contains AA, but Akebiae Caulis does not. It is not easy to distinguish the two by appearance alone. In addition, other TCMs, including Clematidis Armandii Caulis, are often misused as Akebiae Caulis, which may also have potential issues. This study utilized the two different methods of NGS and qPCR for detection. The NGS and qPCR methods differed significantly in the detection of Akebiae Caulis and adulterants. The main difference is that only Clematidis Armandii Caulis was detected in the compound by qPCR. However, NGS found that Clematis peterae was used in the commercial samples (Table 3). This raised the question of why qPCR and NGS yielded different results. We checked the ITS2 sequences of Clematis peterae and Clematidis Armandii Caulis and found a high homology of ITS2 sequences between the two species. The primer pairs of Clematidis Armandii Caulis would not mark off both of them (Figure 3), which led to the difference in identification results between qPCR and NGS. An identical positive correlation was observed between qPCR cycle threshold (Ct) values and the Clematidis Armandii Caulis NGS reads obtained from samples of LDXGW137-139. Samples with lower qPCR Ct values generally yielded more sequencing reads.
Table 3
| Herbal material | Species | LDXGW137 | LDXGW138 | LDXGW139 | |||||
|---|---|---|---|---|---|---|---|---|---|
| qPCR | NGS | qPCR | NGS | qPCR | NGS | ||||
| Mu Tong | Akebia quinata | ||||||||
| Akebia trifoliata | |||||||||
| Akebia trifoliata var. australis | |||||||||
| Chuan Mutong | Clematis armandii | √ | √ | √ | √ | ||||
| Clematis montana | √ | √ | √ | √ | |||||
| Guan Mutong | Aristolochia manshuriensis | ||||||||
| Tie-Xian Lian | Clematis peterae | √ | √ | √ | |||||
qPCR, quantitative real-time polymerase chain reaction; LDXGW, Longdan Xiegan Wan; ITS2, internal transcribed spacer 2; NGS, next-generation sequencing.

Discussion
The NGS results of this study show that the advantage of NGS lies in the discovery of unknown added components or unknown fakes. The NGS method can provide identification of elements based on any amplifiable DNA molecular tag sequence. Nevertheless, the processing or manufacture of compound Chinese medicinal materials will lead to the degradation of DNA in the restorative materials (2,22), resulting in no molecular tags being amplified and the occurrence of false negatives. On the contrary, shorter PCR primers can solve the false negative detection due to DNA degradation (23). Due to the difference in technical principles, NGS is more expensive. It takes longer to detect DNA molecular tags than qPCR technology, but NGS becomes more cost-effective as more samples are processed (24,25). As NGS is an emerging technology, and its sequencing cost is decreasing year by year, higher sample throughput and greater affordability will be possible in the future. Therefore, in the long run, NGS will become more cost-effective, provide more data at a lower cost, and approach the detection sensitivity of qPCR in compounds without processed medicinal materials. Each authentication method has limitations. The NGS methods have different sequencing error rates, depending on the platform: about 1% for Ion-TorrentTM Personal Genome Machines (PGM; Thermo Fisher, Waltham, MA, USA), about 1% for Illumina platforms (Illumina, San Diego, CA, USA), and about 15% for Pacific Biosciences (Menlo Park, CA, USA) (26). Sequencing errors appear similar to those of DNA mutations and can complicate accurate species identification. To identify TCM compound ingredients, NGS technology and qPCR technology should complement each other, not replace. The NGS approach can quickly scan the components in the sample and find unknown added or faulty features. Fluorescence qPCR achieves the purpose of fast, fixed-point detection with its quick, simple, and economic characteristics.
In the 2015 edition of the Chinese Pharmacopoeia, there are 15 TCM compounds containing Mu Tong, but there is no description of the identification method of Mu Tong in the compound form. AAs exist in many plants of the Aristolochiaceae family, which pose a severe threat to human life and health (27). The “Longdan Xiegan Pill Incident” in the 1990s resulted in a painful TCM safety incident due to the lack of effective means to detect Mu Tong medicinal materials (28). There have been many reports of instances where Chuan Mutong has also replaced Mu Tong (29). If there is a continued lack of effective detection methods and methods, potential danger will remain a threat to the safety of TCM. This study is a supplement for detecting compound Mu Tong in TCM products.
Acknowledgments
Funding: This work was supported by the Special Project of the National Clinical Research Base of Traditional Chinese Medicine (No. 2021JDZX2090).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-2415/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-2415/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-2415/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sankararaman S, Velayuthan S, Chen Y, et al. Role of Traditional Chinese Herbal Medicines in Functional Gastrointestinal and Motility Disorders. Curr Gastroenterol Rep 2022;24:43-51. [Crossref] [PubMed]
- de Boer HJ, Ichim MC, Newmaster SG. Correction to: DNA Barcoding and Pharmacovigilance of Herbal Medicines. Drug Saf 2021;44:397. [Crossref] [PubMed]
- Chen YG, He XL, Huang JH, et al. Impacts of heavy metals and medicinal crops on ecological systems, environmental pollution, cultivation, and production processes in China. Ecotoxicol Environ Saf 2021;219:112336. [Crossref] [PubMed]
- Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China (2015 Edition). Beijing: China Medical Science and Technology Press, 2015.
- Lu G, Qiao J, Wang L, et al. An integrated study of Violae Herba (Viola philippica) and five adulterants by morphology, chemical compositions and chloroplast genomes: insights into its certified plant origin. Chin Med 2022;17:32. [Crossref] [PubMed]
- Raclariu AC, Paltinean R, Vlase L, et al. Comparative authentication of Hypericum perforatum herbal products using DNA metabarcoding, TLC and HPLC-MS. Sci Rep 2017;7:1291. [Crossref] [PubMed]
- Wang N, Xing RR, Zhou MY, et al. Application of DNA barcoding and metabarcoding for species identification in salmon products. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 2021;38:754-68. [Crossref] [PubMed]
- Li Q, Sun Y, Guo H, et al. Quality control of the traditional Chinese medicine Ruyi jinhuang powder based on high-throughput sequencing and real-time PCR. Sci Rep 2018;8:8261. [Crossref] [PubMed]
- Nichols RV, Curd E, Heintzman PD, et al. Targeted Amplification and Sequencing of Ancient Environmental and Sedimentary DNA. Methods Mol Biol 2019;1963:149-61. [Crossref] [PubMed]
- Ragupathy S, Faller AC, Shanmughanandhan D, et al. Exploring DNA quantity and quality from raw materials to botanical extracts. Heliyon 2019;5:e01935. [Crossref] [PubMed]
- Li W, Hou X, Xu C, et al. Validating eDNA measurements of the richness and abundance of anurans at a large scale. J Anim Ecol 2021;90:1466-79. [Crossref] [PubMed]
- Li X, Xia Y, Li G, et al. Traditional uses, phytochemistry, pharmacology, and toxicology of Akebiae Caulis and its synonyms: A review. J Ethnopharmacol 2021;277:114245. [Crossref] [PubMed]
- Wu L, Wang B, Zhao M, et al. Rapid Identification of Officinal Akebiae Caulis and Its Toxic Adulterant Aristolochiae Manshuriensis Caulis (Aristolochia manshuriensis) by Loop-Mediated Isothermal Amplification. Front Plant Sci 2016;7:887. [Crossref] [PubMed]
- Gao SS, Li JR, Wu FB, et al. Development of FTIR fingerprint for identification of armand clematis stem (Chuanmutong) and related herbs. Zhongguo Zhong Yao Za Zhi 2016;41:1485-92. [PubMed]
- Gao Y, Xiao XH, Zhu XX, et al. Study and opinion on toxicity of aristolochic acid. Zhongguo Zhong Yao Za Zhi 2017;42:4049-53. [PubMed]
- Yang B, Xie Y, Guo M, et al. Nephrotoxicity and Chinese Herbal Medicine. Clin J Am Soc Nephrol 2018;13:1605-11. [Crossref] [PubMed]
- Intharuksa A, Sasaki Y, Ando H, et al. The combination of ITS2 and psbA-trnH region is powerful DNA barcode markers for authentication of medicinal Terminalia plants from Thailand. J Nat Med 2020;74:282-93. [Crossref] [PubMed]
- Uluar O, Çiplak B. Evolution, characterization and phylogenetic utility of ITS2 gene in Orthoptera and some Polyneoptera: Highly variable at the order level and highly conserved at the species level. Zootaxa 2020;4780:zootaxa.4780.1.2.
- Fujii T, Mori T, Tatsuo Y, et al. Identification of Valeriana fauriei and other Eurasian medicinal valerian by DNA polymorphisms in psbA-trnH intergenic spacer sequences in chloroplast DNA. J Nat Med 2021;75:699-706. [Crossref] [PubMed]
- Nithaniyal S, Majumder S, Umapathy S, et al. Forensic application of DNA barcoding in the identification of commonly occurring poisonous plants. J Forensic Leg Med 2021;78:102126. [Crossref] [PubMed]
- Xiong C, Sun W, Li J, et al. Identifying the Species of Seeds in Traditional Chinese Medicine Using DNA Barcoding. Front Pharmacol 2018;9:701. [Crossref] [PubMed]
- Song M, Dong GQ, Zhang YQ, et al. Identification of processed Chinese medicinal materials using DNA mini-barcoding. Chin J Nat Med 2017;15:481-6. [Crossref] [PubMed]
- Lacoursière-Roussel A, Dubois Y, Normandeau E, et al. Improving herpetological surveys in eastern North America using the environmental DNA method. Genome 2016;59:991-1007. [Crossref] [PubMed]
- Yoshinaga Y, Daum C, He G, et al. Genome Sequencing. Methods Mol Biol 2018;1775:37-52. [Crossref] [PubMed]
- Bacquet J, Stojkovic T, Boyer A, et al. Molecular diagnosis of inherited peripheral neuropathies by targeted next-generation sequencing: molecular spectrum delineation. BMJ Open 2018;8:e021632. [Crossref] [PubMed]
- Mustafa AE, Faquih T, Baz B, et al. Validation of Ion TorrentTM Inherited Disease Panel with the PGMTM Sequencing Platform for Rapid and Comprehensive Mutation Detection. Genes (Basel) 2018;9:267. [Crossref] [PubMed]
- Rebhan K, Ertl IE, Shariat SF, et al. Aristolochic acid and its effect on different cancers in uro-oncology. Curr Opin Urol 2020;30:689-95. [Crossref] [PubMed]
- Nortier JL, Martinez MC, Schmeiser HH, et al. Urothelial carcinoma associated with the use of a Chinese herb (Aristolochia fangchi) N Engl J Med 2000;342:1686-92. [Crossref] [PubMed]
- Huang D, Jin L, Lin L, et al. Investigation of Akebiae Caulis and Clematidis Armandii Caulis. Chinese Journal of Information on Traditional Chinese Medicine 2017;1-4.
(English Language Editor: J. Jones)


