
Identification of the key genes in chronic obstructive pulmonary disease by weighted gene co-expression network analysis
Introduction
Chronic obstructive pulmonary disease (COPD) is characterized by progressive, irreversible limited airflow. Smoking is the most important risk factor, so smoking cessation is an efficient intervention to protect against COPD. However, the pathophysiology of COPD is complex and there is a lack of highly specific therapies. The main pathological disease manifestations include alveolar destruction, airway inflammation, apoptosis, microvasculature remodeling, and microvasculature proliferation (1). The Chinese Adult Lung Health study reported a COPD prevalence rate in adults aged over 40 years in China of 13.7%, accounting for approximately 100 million individuals (2). By 2030, the World Health Organization has estimated that approximately 5.4 million people will die from COPD and associated conditions.
Several strategies can limit COPD progression, and have clinical efficacy (e.g., inhaled corticosteroids, long-acting muscarinic antagonists, and long-acting β2 agonists), but such treatments only slow disease progression, and ultimately fail to reverse disease progression (3). The precise pathogenesis underlying COPD remains unclear, so it is important to investigate and identify novel therapeutic targets.
It has not only been reported that COPD is regulated by multiple genes, but also there is some degree of familial clustering (4). Gene chip technologies are being increasingly used in scientific and clinical research (e.g., gene chip and RNA sequencing strategies). In 2008, Wang et al. investigated the gene expression profiles in lung specimens from 48 COPD patients and used candidate genes, such as transforming growth factor-β and serpin family E member 2, in their screening approach (5). Sun et al. explored differentially expressed genes (DEGs) in COPD and non-COPD patients using bioinformatics analyses and screened autophagy-related genes for validation, because they considered these genes were clinically significant and exerted some pathogenic effects in COPD (6). Thus, an in-depth exploration of public datasets and application of multiple validation models identifying the genetic variants related to COPD could be used to predict disease prognosis and generate individualized treatments for COPD.
To this end, we used 3 expression profiles from the online Gene Expression Omnibus (GEO) database and identified 326 downregulated DEGs and 503 upregulated DEGs. We conducted Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses to identify the potential function of these DEGs. Subsequently, two phenotypes of COPD were combined for key module screening. We performed a module partition analysis using weighted gene co-expression network analysis (WGCNA) to further assess the significant modules. And five different algorithms were used to select the hub genes within the modules, which led to more accurate gene screening results. We present the following article in accordance with the STREGA reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-2523/rc).
Methods
Clinical sample
We selected four paraneoplastic tissue samples from COPD patients and non-COPD patients in The First Affiliated Hospital of Jinan University, respectively. All patients with COPD had a history of smoking ranging from 10 to 30 years, and none of the patients without COPD had a history of smoking. The study was approved by Ethics Committee of Jinan University (No. KY-2021-051). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all the patients.
COPD-related gene datasets and microarray data
Species was set as Homo sapiens and the specimen type was set as lung tissue. Three COPD gene expression profiles, comprising GSE38974, GSE106986, and GSE76925, were explored in the GEO database (https://www.ncbi.nlm.nih.gov/geo/). Our dataset included 148 COPD samples and 54 controls. An additional profile GSE69818 included 70 COPD samples, categorized by the presence or absence of emphysema and Global Initiative for Chronic Obstructive Lung Disease (GOLD) stage.
DEG analysis of COPD-related genes
We used the Limma R package to investigate DEGs in each GEO dataset. DEGs with |log2fold-change (FC)| >0.5 and adjusted P<0.05 were considered the cut-off criteria.
GO and KEGG pathway enrichment analyses
To analyze the main biological functions of the DEGs, GO and KEGG pathway analyses were performed using clusterProfiler in R. GO functional categories consisted of molecular function (MF), biological process (BP), and cellular component (CC). KEGG analyses identified significant DEG pathways. A corrected P value <0.05 was considered statistically significant.
Protein-protein interaction (PPI)
Those DEGs was applied to PPI analysis using a Search Tool called STRING (https://cn.string-db.org/). Then imported the interaction data into Cytoscape, five kinds of algorithms were selected for analysis.
WGCNA of DEGs
We used the WGCNA package in R (https://cran.r-project.org/package=WGCNA) to perform WGCNA analyses based on GSE69818, which included the gene expression data of 70 patients with COPD. Soft thresholding powers were determined using the pick Soft Threshold function in WGCNA. The soft thresholding power parameter β was set at β=6. A topological overlap matrix-based dissimilarity measure was used for the hierarchical clustering of genes to divide similar genes into modules. Module membership (MM) represented correlations between the gene expression values across samples and the module eigenprotein (ME). Correlations between gene expression profiles versus the ME were used to quantify how close the genes were to a given module. Modules were identified and designated by an assigned color.
Cell culture, cigarette smoke extract (CSE) preparation, and treatments
Human 16HBe and THP-1 cell lines obtained from Sun Yat-sen University were cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum and penicillin-streptomycin (Gibco) at 37 ℃ in 5% CO2. Cigarettes (Hong Shuang Xi manufactured by Guangdong China Tobacco Industry Co., Ltd.) used in this study emitted 11 mg tar, 1.0 mg nicotine, and 13 mg CO per cigarette. The cigarettes were connected to the gas sampling, bringing the smoke in contact with cell culture medium by repeated aspiration using a 50-mL syringe. The optical density (OD) value obtained at a wavelength of 490 nm equivalent to 0.25 was defined as 100%. Cells were exposed to 5% CSE for 72 h.
RNA extraction and quantitative polymerase chain reaction (qPCR)
RNA was extracted using the TRIzol reagent (Invitrogen, San Diego, CA, USA) according to the manufacturer’s instructions. Total RNA was reverse transcribed using a reverse transcription kit (Ericbio, China). SYBR green (iQ SYBR green supermix, BioRad) quantitative polymerase chain reaction was performed using CFX instrument (BioRad, Hercules, CA, USA). Reactions contained 10 µL SYBR Green 1×, 1 µL cDNA, and 0.4 µL reverse primers. Relative fold change (FC) was determined from cycle threshold values using the 2−ΔΔCt method.
Statistical analysis
Statistical analysis was performed using R language software (version 4.0.2, https://www.r-project.org).
Results
DEG identification
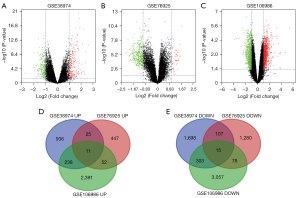
We selected the genetic data for 148 COPD and 54 non-COPD lung tissues from the GSE38974, GSE106986, and GSE76925 datasets. DEG analysis was performed for each dataset in GEO2 R. A total of 1,210 upregulated genes and 2,123 downregulated genes were screened from GSE38974; 535 upregulated genes and 1,480 downregulated genes from GSE106986; and 2,692 upregulated genes and 3,453 downregulated genes from GSE76925. There were 326 upregulated genes (log2FC >1 and adjusted P<0.05) and 503 common downregulated genes (log2FC <−1 and adjusted P<0.05) in at least 2 datasets. The datasets are visually represented by volcano charts (Figure 1A-1C). Overlaps in the DEG data are illustrated by Venn diagram (Figure 1D,1E).
DEG GO and KEGG analyses
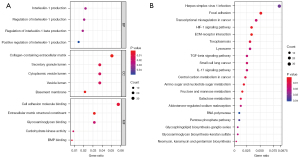
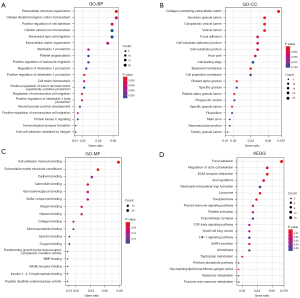
To explore the DEGs biological functions, GO and KEGG term enrichment analyses of the identified 326 upregulated genes and 503 downregulated genes were performed using the clusterProfiler package in R. For BP terms, DEGs were significantly enriched for interleukin-1 (IL-1) production, regulation of IL-1 production, regulation of IL-1β production, and positive regulation of IL-1 production. DEGs were significantly enriched in the “inflammatory response” pathways. For CC terms, DEGs were significantly enriched in the extracellular regions and vesicle lumen, such as collagen-containing extracellular matrix (ECM), basement membranes, secretory granule lumen, cytoplasmic vesicle lumen, and vesicle lumen. For MF terms, DEGs were mainly enriched in the ECM structural constituents, cell adhesion molecule binding, bone morphogenetic protein (BMP) binding, carbohydrate kinase activity, and glycosaminoglycan binding (Figure 2A). KEGG pathway analyses indicated the most significantly enriched pathways for DEGs were ECM-receptor interactions, the hypoxia-inducible factor-1 (HIF-1) signaling pathway, and focal adhesion. These signaling pathways were related to the inflammatory reaction processes and alveolar and airway remodeling (Figure 2B).
WGCNA and key module visualization
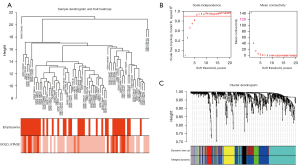
The gene co-expression network established by WGCNA in R. A total of 70 COPD lung tissue samples were clustered by Pearson’s correlation and average linkage algorithms (Figure 3A). β=6 was chosen as the soft-thresholding parameter to ensure a scale-free network. Also, the fitting degree of the scale-free topological model was set at 9.0 (Figure 3B). Then, the ME values were analyzed for correlations with tissue or disease traits. In total, 7 expression modules were identified using average linkage hierarchical clustering (Figure 3C). In terms of COPD phenotypes, the top 4 correlated modules were yellow (86 DEGs), black (42 DEGs), green (50 DEGs), and blue (228 DEGs), which showed high R2 values and low P values. A network diagram was generated according to the weight value of each module and hub genes in each identified functional module (Figure 4).
“Hub” genes and module identification
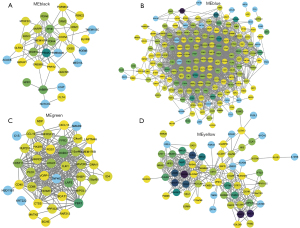
Cytoscape was used to visualize the co-expression network of each module to screen for the key genes. The node and edge represented the gene and weight, respectively, between 2 genes. The relationship between MM and gene significance (GS) showed that 4 gene modules had significant correlations with COPD (Figure 5).
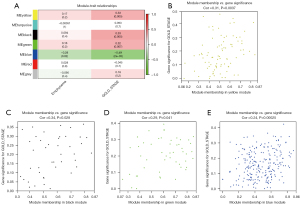
A scatterplot of GS was plotted to define module trait associations between gene expression and clinical characteristics of COPD patients; each module showed positively correlated GS and MM values (P<0.05). Yellow, black, and green modules were positively correlated with the GOLD stage, especially the black module (correlation coefficient; 0.34, P=0.028). The green module was positively correlated with the emphysematous phenotype (correlation coefficient: 0.24, P=0.00025).
Next, we selected genes from these 4 modules to further analyze enrichment levels. In the BP functional category, DEGs were enriched in the extracellular structure organization, positive regulation of IL-1 production, regulation of mononuclear cell migration, positive regulation of cell adhesion, and positive regulation of IL-1β production (P<0.01) (Figure 6A). For CC, DEGs were specifically focused on the basement membrane, collagen-containing ECM, secretory granule lumen, cytoplasmic vesicle lumen, and vesicle lumen (P<0.01) (Figure 6B). For MF, DEGs were primarily assembled into the ECM structural constituents, cell adhesion molecule binding, calmodulin binding, integrin binding, and glycosaminoglycan binding (P<0.01) (Figure 6C). In the KEGG analyses, DEGs were mostly enriched in focal adhesion, regulation of actin cytoskeleton, ECM-receptor interaction, axon guidance, and neutrophil extracellular trap formation (Figure 6D).
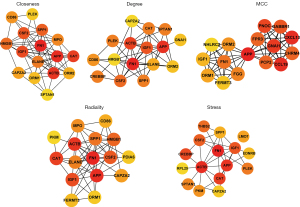
Identification of hub genes using the PPI network
A PPI network of the DEGs in the 4 significant modules was selected for PPI network analysis in STRING (Figure 7). After ranking by several indices, such as closeness, degree, MCC (maximal clique centrality), radiality, and stress, the top 16 genes in these 5 indices were selected from the PPI network using the plug-in, cytoHubba. Considering the differences in the output of different algorithms, the following genes that co-existed in at least 4 algorithms were selected as the final hub genes: APP, FN1, IGF1, ACTB, CAPZA2, SPP1, CAT, and CSF2 (Figure 8).

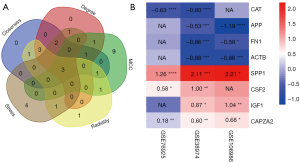
Validation of gene transcription levels
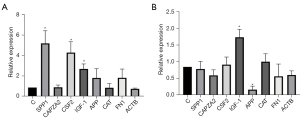
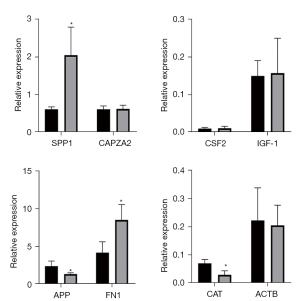
A Venn diagram of the hub genes from the 5different algorithms was constructed. Eight genes in the overlapping regions of at least 4 algorithms were selected for further analysis (Figure 9A). Additionally, expression heatmaps of hub genes in the GSE76925, GSE38974, and GSE106986 datasets are shown in Figure 9B. The CSE-stimulated 16HBE cells were found to highly express SPP1, CSF, and IGF1. In addition, IGF1 levels were increased and APP levels were decreased in CSE-stimulated THP-1 cells (Figure 10). SPP1 and FN1 had increased expression levels in COPD lung tissues, the opposite held for APP and CAT (Figure 11).
Discussion
Although the pathophysiology of COPD is complex and there is a lack of highly specific drugs to treat this disease, smoking is the most important risk factor for the disease. In developed countries, the populations of individuals who smoke represent approximately 70% of global smokers. In China, more than 300 million people smoke; in 2018, the smoking prevalence was 50.5% among men and 26.6% in individuals aged >15 years (7). Therefore, identifying specific therapeutic targets of smoking-induced pathogenic mechanisms could be beneficial in preventing and treating COPD.
Hub genes are central to disease-associated protein regulatory networks, so associated genetic studies may provide vital clues to the pathogenesis of COPD. In this study, gene expression profile data from the GEO database were analyzed for DEGs implicated in COPD.
We selected 3 datasets, which included 148 COPD and 54 non-COPD lung tissue profiles, and we identified 326 upregulated genes and 503 downregulated genes. GO analyses showed that DEGs were enriched in the inflammatory reaction processes and alveolar and airway remodeling. KEGG analysis showed that DEGs were enriched in focal adhesion, regulation of actin cytoskeleton, and ECM-receptor interaction. Chronic inflammation in COPD and acute exacerbation of COPD (AECOPD) are related to altered homeostasis of ECM molecules in the lungs (8,9). Cells require attachment to the ECM for survival, and detachment induces apoptosis of many types of cells (10). The actin cytoskeleton may mediate smooth muscle binding with the ECM to form focal adhesions, which is important in airway remodeling (11). In the GSE69818 dataset, 782 DEGs were determined, which were then subjected to WGCNA to identify the key functional modules. Next, we carried out GO and KEGG enrichment analyses for the genes in the key modules, and we found that DEGs were also enriched in the inflammatory reaction processes and alveolar and airway remodeling. Furthermore, PPI network analysis of these DEGs with 5 algorithms was also performed and finally we obtained 8 hub genes: APP, FN1, IGF1, ACTB, CAPZA2, SPP1, CAT, and CSF2.
A portion of these hub genes have been verified to be associated with COPD, but the other is unknown. CAT is an H2O2 scavenger, and a key enzyme of the biological defense system (12). In patients with smoking-related COPD, the expression of CAT in the bronchiolar epithelium decreases dramatically (13). FN1 is an ECM glycoprotein (14), and in this study its expression was decreased by CSE treatment, which might promote hormone resistance in COPD (15). CAPZA2 is one of the components of the cytoskeleton (16), but the association between CAPZA2 and COPD is poorly understood. The same holds true for ACTB.
The CSE-stimulated 16HBE cells were found to highly express SPP1, CSF, and IGF1. In addition, IGF1 levels were increased and APP levels were decreased in CSE-stimulated THP-1 cells. CSF2, also known as granulocyte-macrophage colony-stimulating factor (GM-CSF), is a major survival and activating factor for macrophages and neutrophils in the lung (17). Cigarette smoking increases the expression of pulmonary GM-CSF and granulocytes, resulting in pulmonary inflammation (18). Shen et al. found that carriers of the CSF2 117Ile allele had a 2.4-fold higher risk of COPD than the wild-type (Thr/Thr) carriers when exposed to indoor air pollution (19).
SPP1, or osteopontin, participates in bone metabolism, immune responses, and cancer metastasis (20). Papaporfyriou et al. showed that SPP1 sputum levels were higher in patients with COPD than in asymptomatic smokers and non-smokers (21). Cigarette smoke-induced inflammatory cytokine secretion and inflammatory cell infiltration play key roles in COPD. SPP1 is a chemokine that regulates immune cell differentiation and proliferation (22). Shan et al. showed that exposure of dendritic cells to CSE induced SPP1 mRNA expression and stimulated Th17 cell differentiation, thus mediating interleukin-17A (IL-17A) driven inflammation, and ultimately resulting in emphysema (23). SPP1 knockdown attenuated CSE-induced immune cell infiltration, IL-17A production, and alveolar destruction. Thus, SPP1 appears to be involved in the pathogenesis of emphysema by regulating innate and adaptive immune responses.
SPP1 is closely associated with the phosphatidylinositol 3-kinase (PI3K)-protein kinase B (AKT) signaling pathway, and activation of the Toll-like receptor (TLR) signaling pathway (24,25). PI3K signaling regulates growth, proliferation, survival, metabolism, and angiogenesis in vitro and in vivo. Activated AKT regulates cell functions by phosphorylating various enzymes, transcription factors, and kinases; thus, aberrant AKT activity may affect these fundamental processes (26,27). SPP1 could regulate inflammatory cell activation, inflammatory mediator release, and airway remodeling in COPD by intervening in the PI3K/AKT signaling pathway.
Similarly, it has been reported that TLRs are important in COPD pathogenesis (28). These receptors belong to the pattern recognition receptor (PRR) family. Cigarette smoke stimulates immune competent cells in the respiratory tract, potentially activating TLRs (29). This activation may be mediated by damage-associated molecular patterns (DAMPs) and high mobility group box-1 (HMGB1), heat shock protein 60 (HSP60), HSP70, and β-defensin. We therefore speculate that SPP1, which is involved in the TLR and PI3K/AKT mechanisms, is stimulated by cigarette smoke, which in turn leads to COPD progression.
In our study, the APP transcription levels in THP-1 cells were decreased after CSE exposure for 72 h, which was consistent with our bioinformatics data. APP is a protein-coding gene related to cerebral amyloid angiopathy and Alzheimer’s disease (30). It regulates axonal growth, neuronal adhesion, and axonogenesis, and participates in cell movement, proliferation, and transcription (31). However, APP has not been thoroughly investigated in pulmonary disease etiology. Spitzer et al. have reported that APP is hydrolyzed to β-amyloid (Aβ) in macrophage lineages, and stimulated pro-inflammatory cytokine secretion from macrophages (32). APP also regulates tumor necrosis factor-α (TNF-α), IL-6, and IL-10 secretion from human monocyte-derived macrophages. APP knockdown in macrophages decreased TNF-α and IL-6 levels and increased the IL-10 level (32). However, IL-10 secretion was diminished during lipopolysaccharide (LPS)-induced inflammation. Puig et al. compared C57BL6 wild-type and APP−/− mice, and found that the migratory capacity of peritoneal macrophages and cytokine secretion levels were reduced in the latter (33). In an infectious meningitis study, Kumar et al. reported that APP−/− mice had lower survival rates, and its overexpression had the opposite effect (34). Thus, APP appears to play an anti-infection role in macrophages. In our study, we identified decreased APP transcription levels in macrophages after exposure to CSE. Theoretically, smoking induces decreased immune responses in the respiratory tract, which may explain why smokers are more likely to acquire respiratory infections.
The SPP1 and APP levels correlate with lung fibrosis. Pardo et al. have reported that SPP1 was significantly increased in the bronchoalveolar lavage fluid of patients with idiopathic pulmonary fibrosis (IPF) (35). SPP1 promotes the proliferation and migration of primary human lung epithelial cells. Also, patients with IPF have elevated serum SPP1 levels. Moreover, patients with an acute exacerbation had higher SPP1 levels than patients without an acute exacerbation (36). Several studies have suggested that SPP1 is not merely a biomarker of lung fibrosis, but may also promote the progression of lung fibrosis (37,38). Thus, SPP1 potentially represents a viable therapeutic target for lung fibrosis. APP promotes pulmonary fibrosis by regulating macrophage phenotypes. Monocyte-derived macrophages are polarized to pro-inflammatory and pro-fibrotic phenotypes (39). APP in the induced pluripotent stem cell secretome caused a switch from a pro-fibrotic to a pro-inflammatory phenotype. After APP was scavenged by a specific antibody, the pro-fibrotic phenotype in the macrophage subsets was increased (40). IL-10 is an important anti-inflammatory cytokine and anti-fibrotic factor, LPS-induced secretion of IL-10 was also suppressed in APP knockout macrophages (32). Lung fibrosis can develop in some patients with advanced COPD, SPP1 and APP may contribute to this change.
IGF is a protein with a structure and function similar to insulin and is localized to the interstitial lung tissue, where it is involved in lung development and anabolism. The IGF system plays an important role in lung development (41,42). The pathology of bronchopulmonary dysplasia includes thickened interstitial tissue in the lungs and enhanced cell densities, accompanied by the enhanced expression of IGF1 (43). Knockout of the IGF1 receptor gene causes pulmonary dysplasia (44). COPD patients have low serum levels of IGF1; in a severe COPD group there was a significant difference compared with the mild to moderate COPD group (45). IGF1 is also considered to be a key regulator of muscle quality, and muscle dysfunction is one of the most common systemic manifestations of COPD. The presence of IGF-1 promotes the formation of myotubes and the expression of muscle-specific protein. Moreover, it can prevent the muscle atrophy induced by glucocorticoid treatment (46). Conversely, a reduction in IGF1 levels accelerates muscle wasting. Prenatal tobacco exposure increased the mRNA level of IGF1 in offspring mice, and caused pulmonary dysplasia (47). Nicotine, a component of cigarette smoke, can induce the macrophage phenotype switch towards M2. M2 macrophages promote tumorigenesis via the secretion of IGF1 (48). The serum IGF1 level in smokers is positively linked to lung cancer risk (49). This is consistent with our bioinformatics analysis and qPCR results, which illustrates the complex regulatory relationship between IGF1 and cigarette smoke-induced COPD. Moreover, IGF1 might play different roles in different cell types.
However, our study has some limitations. Since the data did not contain prognostic information, it was not possible to analyze the relationship between these pivotal genes and patient prognosis. And our lung tissues were from lung cancer patients, which inevitably interfered with our prognostic judgments.
Conclusions
In conclusion, we found 8 hub genes by bioinformatics analysis, and finally identified 4 significant DEGs validated through qPCR: SPP1, CSF, IGF1, and APP. However, the limitation of this study is that inter-ethnic differences and cellular diversity were not considered. However, our results have provided insights into the pathophysiological mechanisms of COPD.
Acknowledgments
Funding: This work was supported by the Guangzhou Science, Technology Planning Program (No. 202102010064).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-2523/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-2523/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-2523/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of Jinan University (No. KY-2021-051). Informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Page C, O'Shaughnessy B, Barnes P. Pathogenesis of COPD and Asthma. Handb Exp Pharmacol 2017;237:1-21. [Crossref] [PubMed]
- Wang C, Xu J, Yang L, et al. Prevalence and risk factors of chronic obstructive pulmonary disease in China (the China Pulmonary Health [CPH] study): a national cross-sectional study. Lancet 2018;391:1706-17. [Crossref] [PubMed]
- Wheaton AG, Liu Y, Croft JB, et al. Chronic Obstructive Pulmonary Disease and Smoking Status - United States, 2017. MMWR Morb Mortal Wkly Rep 2019;68:533-8. [Crossref] [PubMed]
- Silverman EK. Genetics of COPD. Annu Rev Physiol 2020;82:413-31. [Crossref] [PubMed]
- Wang IM, Stepaniants S, Boie Y, et al. Gene expression profiling in patients with chronic obstructive pulmonary disease and lung cancer. Am J Respir Crit Care Med 2008;177:402-11. [Crossref] [PubMed]
- Sun S, Shen Y, Wang J, et al. Identification and Validation of Autophagy-Related Genes in Chronic Obstructive Pulmonary Disease. Int J Chron Obstruct Pulmon Dis 2021;16:67-78. [Crossref] [PubMed]
- Global Adult Tobacco Survey (GATS) China 2018 Report. Beijing: People’s Medical Publishing House, 2019.
- Postma DS, Timens W. Remodeling in asthma and chronic obstructive pulmonary disease. Proc Am Thorac Soc 2006;3:434-9. [Crossref] [PubMed]
- Barnes PJ. Mediators of chronic obstructive pulmonary disease. Pharmacol Rev 2004;56:515-48. [Crossref] [PubMed]
- Hui AY, Meens JA, Schick C, et al. Src and FAK mediate cell-matrix adhesion-dependent activation of Met during transformation of breast epithelial cells. J Cell Biochem 2009;107:1168-81. [Crossref] [PubMed]
- Tang DD, Gerlach BD. The roles and regulation of the actin cytoskeleton, intermediate filaments and microtubules in smooth muscle cell migration. Respir Res 2017;18:54. [Crossref] [PubMed]
- Galasso M, Gambino S, Romanelli MG, et al. Browsing the oldest antioxidant enzyme: catalase and its multiple regulation in cancer. Free Radic Biol Med 2021;172:264-72. [Crossref] [PubMed]
- Betsuyaku T, Fuke S, Inomata T, et al. Bronchiolar epithelial catalase is diminished in smokers with mild COPD. Eur Respir J 2013;42:42-53. [Crossref] [PubMed]
- Goossens K, Van Soom A, Van Zeveren A, et al. Quantification of fibronectin 1 (FN1) splice variants, including two novel ones, and analysis of integrins as candidate FN1 receptors in bovine preimplantation embryos. BMC Dev Biol 2009;9:1. [Crossref] [PubMed]
- Pei G, Ma N, Chen F, et al. Screening and Identification of Hub Genes in the Corticosteroid Resistance Network in Human Airway Epithelial Cells via Microarray Analysis. Front Pharmacol 2021;12:672065. [Crossref] [PubMed]
- Huang Y, Mao X, van Jaarsveld RH, et al. Variants in CAPZA2, a member of an F-actin capping complex, cause intellectual disability and developmental delay. Hum Mol Genet 2020;29:1537-46. [Crossref] [PubMed]
- Vlahos R, Bozinovski S, Hamilton JA, et al. Therapeutic potential of treating chronic obstructive pulmonary disease (COPD) by neutralising granulocyte macrophage-colony stimulating factor (GM-CSF). Pharmacol Ther 2006;112:106-15. [Crossref] [PubMed]
- Jamal Jameel K, Gallert WJ, Yanik SD, et al. Biomarkers for Comorbidities Modulate the Activity of T-Cells in COPD. Int J Mol Sci 2021;22:7187. [Crossref] [PubMed]
- Shen M, Vermeulen R, Chapman RS, et al. A report of cytokine polymorphisms and COPD risk in Xuan Wei, China. Int J Hyg Environ Health 2008;211:352-6. [Crossref] [PubMed]
- Wang KX, Denhardt DT. Osteopontin: role in immune regulation and stress responses. Cytokine Growth Factor Rev 2008;19:333-45. [Crossref] [PubMed]
- Papaporfyriou A, Loukides S, Kostikas K, et al. Increased levels of osteopontin in sputum supernatant in patients with COPD. Chest 2014;146:951-8. [Crossref] [PubMed]
- O'Regan A. The role of osteopontin in lung disease. Cytokine Growth Factor Rev 2003;14:479-88. [Crossref] [PubMed]
- Shan M, Yuan X, Song LZ, et al. Cigarette smoke induction of osteopontin (SPP1) mediates T(H)17 inflammation in human and experimental emphysema. Sci Transl Med 2012;4:117ra9. [Crossref] [PubMed]
- Dai J, Peng L, Fan K, et al. Osteopontin induces angiogenesis through activation of PI3K/AKT and ERK1/2 in endothelial cells. Oncogene 2009;28:3412-22. [Crossref] [PubMed]
- Salvi V, Scutera S, Rossi S, et al. Dual regulation of osteopontin production by TLR stimulation in dendritic cells. J Leukoc Biol 2013;94:147-58. [Crossref] [PubMed]
- Kok K, Geering B, Vanhaesebroeck B. Regulation of phosphoinositide 3-kinase expression in health and disease. Trends Biochem Sci 2009;34:115-27. [Crossref] [PubMed]
- Bozinovski S, Vlahos R, Hansen M, et al. Akt in the pathogenesis of COPD. Int J Chron Obstruct Pulmon Dis 2006;1:31-8. [Crossref] [PubMed]
- Sidletskaya K, Vitkina T, Denisenko Y. The Role of Toll-Like Receptors 2 and 4 in the Pathogenesis of Chronic Obstructive Pulmonary Disease. Int J Chron Obstruct Pulmon Dis 2020;15:1481-93. [Crossref] [PubMed]
- Olloquequi J, Silva OR. Biomass smoke as a risk factor for chronic obstructive pulmonary disease: effects on innate immunity. Innate Immun 2016;22:373-81. [Crossref] [PubMed]
- Selkoe DJ. Toward a comprehensive theory for Alzheimer's disease. Hypothesis: Alzheimer's disease is caused by the cerebral accumulation and cytotoxicity of amyloid beta-protein. Ann N Y Acad Sci 2000;924:17-25. [Crossref] [PubMed]
- Priller C, Bauer T, Mitteregger G, et al. Synapse formation and function is modulated by the amyloid precursor protein. J Neurosci 2006;26:7212-21. [Crossref] [PubMed]
- Spitzer P, Walter M, Göth C, et al. Pharmacological Inhibition of Amyloidogenic APP Processing and Knock-Down of APP in Primary Human Macrophages Impairs the Secretion of Cytokines. Front Immunol 2020;11:1967. [Crossref] [PubMed]
- Puig KL, Swigost AJ, Zhou X, et al. Amyloid precursor protein expression modulates intestine immune phenotype. J Neuroimmune Pharmacol 2012;7:215-30. [Crossref] [PubMed]
- Kumar DK, Choi SH, Washicosky KJ, et al. Amyloid-β peptide protects against microbial infection in mouse and worm models of Alzheimer's disease. Sci Transl Med 2016;8:340ra72. [Crossref] [PubMed]
- Pardo A, Gibson K, Cisneros J, et al. Up-regulation and profibrotic role of osteopontin in human idiopathic pulmonary fibrosis. PLoS Med 2005;2:e251. [Crossref] [PubMed]
- Gui X, Qiu X, Xie M, et al. Prognostic Value of Serum Osteopontin in Acute Exacerbation of Idiopathic Pulmonary Fibrosis. Biomed Res Int 2020;2020:3424208. [Crossref] [PubMed]
- Kumar A, Elko E, Bruno SR, et al. Inhibition of PDIA3 in club cells attenuates osteopontin production and lung fibrosis. Thorax 2022;77:669-78. [Crossref] [PubMed]
- Hatipoglu OF, Uctepe E, Opoku G, et al. Osteopontin silencing attenuates bleomycin-induced murine pulmonary fibrosis by regulating epithelial-mesenchymal transition. Biomed Pharmacother 2021;139:111633. [Crossref] [PubMed]
- Murray PJ, Wynn TA. Obstacles and opportunities for understanding macrophage polarization. J Leukoc Biol 2011;89:557-63. [Crossref] [PubMed]
- Tamò L, Fytianos K, Caldana F, et al. Interactome Analysis of iPSC Secretome and Its Effect on Macrophages In Vitro. Int J Mol Sci 2021;22:958. [Crossref] [PubMed]
- Meyer KF, Krauss-Etschmann S, Kooistra W, et al. Prenatal exposure to tobacco smoke sex dependently influences methylation and mRNA levels of the Igf axis in lungs of mouse offspring. Am J Physiol Lung Cell Mol Physiol 2017;312:L542-l55. [Crossref] [PubMed]
- Joss-Moore LA, Albertine KH, Lane RH. Epigenetics and the developmental origins of lung disease. Mol Genet Metab 2011;104:61-6. [Crossref] [PubMed]
- Albertine KH, Jones GP, Starcher BC, et al. Chronic lung injury in preterm lambs. Disordered respiratory tract development. Am J Respir Crit Care Med 1999;159:945-58. [Crossref] [PubMed]
- Epaud R, Aubey F, Xu J, et al. Knockout of insulin-like growth factor-1 receptor impairs distal lung morphogenesis. PLoS One 2012;7:e48071. [Crossref] [PubMed]
- Coşkun F, Ege E, Uzaslan E, et al. Evaluation of thyroid hormone levels and somatomedin-C (IGF-1) in patients with chronic obstructive pulmonary disease (COPD) and relation with the severity of the disease. Tuberk Toraks 2009;57:369-75. [PubMed]
- Pansters NA, Langen RC, Wouters EF, et al. Synergistic stimulation of myogenesis by glucocorticoid and IGF-I signaling. J Appl Physiol (1985) 2013;114:1329-39. [Crossref] [PubMed]
- Dehmel S, Nathan P, Bartel S, et al. Intrauterine smoke exposure deregulates lung function, pulmonary transcriptomes, and in particular insulin-like growth factor (IGF)-1 in a sex-specific manner. Sci Rep 2018;8:7547. [Crossref] [PubMed]
- Wu SY, Xing F, Sharma S, et al. Nicotine promotes brain metastasis by polarizing microglia and suppressing innate immune function. J Exp Med 2020;217:e20191131. [Crossref] [PubMed]
- Ho GYF, Zheng SL, Cushman M, et al. Associations of Insulin and IGFBP-3 with Lung Cancer Susceptibility in Current Smokers. J Natl Cancer Inst 2016;108:djw012. [Crossref] [PubMed]
(English Language Editor: K. Brown)


