
Prediction of axillary lymph node metastasis in triple-negative breast cancer by multi-omics analysis and an integrated model
Introduction
Axillary lymph node metastasis (LNM) is the most common method of breast cancer invasion (1). Previously, extensive surgical excisions of axillary lymph nodes (ALNs) were a component of modified radical mastectomy for breast cancer. However, this approach is not beneficial to patients whose dissected ALNs are pathologically nonmetastatic (2,3). Excessive ALN dissection might cause unnecessary postoperative complications, such as lymphedema, local pain, paresthesia, and shoulder stiffness (4-6). Currently, an increasing number of breast cancer patients are diagnosed at an early stage, and more than half of them do not have axillary LNM. Sentinel lymph node biopsy (SLNB) has become the standard surgical approach for invasive breast cancer patients with clinically negative axilla (7). ALNs were exempted from surgical excision if the intraoperative pathological assessment of SLNB was negative. However, SLNB still causes pain to patients and has risks for complications. Therefore, some clinical trials have tried to investigate the exempt surgical excision of SLNB in patients through ultrasound and imaging examinations (8). However, the sensitivity and specificity of these methods could not ensure the accuracy of each case. Therefore, tools are needed to help estimate the risk of LNM more accurately.
Many clinical researchers and clinicians have made unremitting efforts in predicting lymph node (LN) status. Currently, there are several LNM prediction models of breast cancer, and most of them are based on clinicopathology (9). However, compared with other subtypes, these models have poor predictive performance for triple-negative breast cancer (TNBC). Some studies tried to predict LN status utilizing messenger RNA (mRNA) sequencing data, but these models still cannot predict axillary LNM accurately in TNBC (10,11). Therefore, it is urgent to develop a tool for predicting the ALN status of TNBC specifically.
In our research, based on the established multi-omics TNBC cohort in Fudan University Shanghai Cancer Center (FUSCC) (12), we compared differences between LN positive and LN negative cases and found potential LN related markers in multi-omics data, including clinicopathological information, mutation data, somatic copy number alteration (SCNA) data and transcriptomic data. Based on these omics data, we attempted to establish an integrated multi-omics model to predict ALN status in TNBC. We present the following article in accordance with the TRIPOD reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-277/rc).
Methods
Patient cohort
In this study, the cohort was based on our previously published Fudan University Shanghai Cancer Center Triple Negative Breast Cancer (FUSCCTNBC) project (12). A total of 445 patients who underwent surgeries at the Department of Breast Surgery, Fudan University, Shanghai Cancer Center (FUSCC; Shanghai, China), from January 29, 2007, to December 17, 2014, were enrolled in the research.
The inclusion criteria for patients in the research study were as follows: (I) each patient did not have any neoadjuvant therapy before surgery; (II) all patients underwent surgical excision of ALNs; (III) all tumor tissues were confirmed to have invasive breast cancer by histopathological diagnosis; (IV) TNBC was diagnosed with negative immunohistochemical tests showing ER, PR and HER2; and (V) most clinicopathological characteristics were available, including age, tumor type, tumor size, histologic grade, and ALN status.
All data above is available to the public, so the approval of the medical ethics committee board was not necessary. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Data availability
All data used in the study can be viewed and accessed in The National Omics Data Encyclopedia (NODE: OEP000155). All sequence data and microarray data can also be downloaded from the Sequence Read Archive (WES and RNA-seq; SRA: SRP157974) and NCBI Gene Expression Omnibus (OncoScan array; GEO: GSE118527).
Generation of DNA-sequencing data
DNA extracted from 265 tumor tissues and matched white blood cells underwent whole exome sequencing (WES). DNA was fragmented on a Bioruptor Plus sonication system and sequenced on an Illumina HiSeq X TEN platform (Illumina Inc., San Diego, CA, USA). To generate the mutational profile, BWA-mem, Sentieon tools, FastQC, Samtools, VarScan2, TNseq and TNscope were performed in turn. To improve specificity, a panel of normal (PON) samples filtering, processSomatic and somaticFilter tools were used. In addition, mutations in at least two out of three callers (TNseq, TNscope and VarScan2) were included to construct the final somatic mutation compendium.
Generation of somatic copy number variation (SCNV) data
Genome-wide somatic copy number analysis of 385 samples analysis was performed based on the protocol of the OncoScan CNV Assay Kit (Affymetrix, Santa Clara, CA, USA). Cluster intensity values were automatically calculated in an algorithm from DAT files using GeneChip Command Console software (Affymetrix, Inc.). OncoScan Console 1.3 software (Affymetrix, Inc.) and GISTIC2.0 (v2.0.22) were performed to generate peak level and gene level copy number values. The Advanced Scatterometer (ASCAT) algorithm was utilized to adjust copy numbers of genes based on ploidy and purity.
Construction of the expression profile
RNA sequencing was performed on 346 breast cancer tissues and 88 paired normal breast tissues. The RNA library was prepared in the Illumina TruSeq Stranded Total RNA LT sample preparation kit. Then the libraries were sequenced on the Illumina HiSeq X TEN platform (Illumina Inc., San Diego, CA, USA). The TopHat-Cufflinks pipeline (hg19) was used to generate fragments per kilobase of exon per million reads mapped (FPKM) data. To construct the expression profile with relatively accurate values, we removed genes whose FPKMs were not 0 in more than 30% samples before Combat.
Enrichment and pathway analyses
In this study, the differentially expressed mRNAs (DEMs) were analyzed by the package “limma” in R and met the standard of false discovery fate (FDR) <0.05. Gene Ontology (GO) (13) was used to investigate the biological processes of these DEMs. The metabolic pathways were analyzed with Kyoto Encyclopedia of Genes and Genomes (KEGG) (14). GO and KEGG pathway analyses were both performed utilizing the R package “clusterProfiler” (15).
Statistical analysis
We used Student’s t-test and the Wilcoxon test for the comparison of continuous variables and ordered categorical variables, while Pearson’s chi-square test or Fisher’s exact test was employed to compare unordered categorical variables. The methods used for the analyses involving genomic and transcriptomic data were similar to our previous publications. Copy number amplification was defined as a log2 ratio greater than log2(2.5/2), and copy number deletion was defined as a log2 ratio less than log2(1.5/2) (16). All analyses were performed with R software (R version 3.6.2, Vienna, Austria) and IBM SPSS Statistics (R23.0.0.0). Receiver operating characteristic (ROC) curves were generated to distinguish TNBC patients with and without axillary LNM. Predictive accuracy was determined by measuring the area under the ROC curve (AUROC). The least absolute shrinkage and selection operator (LASSO) regression model and stepwise forward regression were used for marker selection. All tests were two sided, and P<0.05 was considered statistically significant.
Results
Patient cohort and study design
A total of 445 TNBC patients derived from the FUSCCTNBC cohort with available information on lymph node status were included in this study. Some of these patients had genomic (WES: n=265, 59.6%; OncoScan: n=385, 86.5%) and transcriptomic (RNA-seq: n=346, 77.8%) data. Among them, 169 patients had positive lymph node metastasis (LNM) (38.0%) and 276 patients had negative LNM (62.0%). These two groups of patients were similar in terms of the distribution and completeness of omics data (Pearson’s chi-square test, P>0.05) and were comparable in terms of clinical characteristics such as age at diagnosis (P=0.341) and histological type (P=0.689) (Figure 1A).
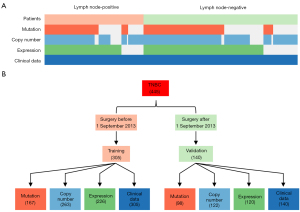
We performed our research mainly in four steps. First, we collected and collated the multi-omics data, and divided the whole cohort into a training set and a validation set. The training set included 305 TNBC patients (68.5%) who underwent surgery before 1 September 2013 at FUSCC, while remaining 140 patients (31.5%) after 1 September 2013 were included in the validation set (Figure 1B). Then, by comparing the clinical information, mutation, copy number and transcriptomic data in the training set, we observed different signatures between LN positive and LN negative cases. In addition, based on these signatures, we developed models to predict lymph node status in TNBC. Finally, by selecting key markers of each omics model, we established a multi-omics prediction model with better performance than models based on individual omics data.
Clinical characteristics of the study population
In total, the study included 445 TNBC patients who underwent axillary surgery, and the clinicopathologic characteristics of the patients in both the training and validation cohorts are shown in Table 1. The baseline clinical and pathological characteristics were generally well balanced between the training and validation cohorts (Pearson’s chi-square test or Fisher’s exact test or Student’s t-test, P>0.05), except for Ki-67 (Fisher’s exact test, P=0.026).
Table 1
| Variable | All patients (n=445), n (%) | Training set (n=305), n (%) | Validation set (n=140), n (%) | P |
|---|---|---|---|---|
| Age, year | 0.386* | |||
| Median | 54 | 54 | 54 | |
| Range | 25–84 | 25–83 | 27–84 | |
| Tumor type | 0.725 | |||
| IDC | 407 (91.5) | 277 (90.8) | 130 (92.9) | |
| ILC | 6 (1.3) | 4 (1.3) | 2 (1.4) | |
| Others | 32 (7.2) | 24 (7.9) | 8 (5.7) | |
| Tumor size, cm | – | – | – | 0.0893* |
| Median | 2.5 | 2.5 | 2.3 | – |
| Median ± SD | [1.4, 3.6] | [1.3, 3.7] | [1.4, 3.2] | – |
| Lymph node metastasis | – | – | – | 0.439 |
| 0 | 276 (62.0) | 186 (61.0) | 90 (64.3) | – |
| 1–3 | 106 (23.8) | 73 (23.9) | 33 (23.6) | – |
| 4–9 | 35 (7.9) | 23 (7.5) | 12 (8.6) | – |
| ≥10 | 28 (6.3) | 23 (7.5) | 5 (3.6) | – |
| Ki-67 | – | – | – | 0.038 |
| <20 | 45 (10.1) | 37 (12.1) | 8 (5.7) | – |
| ≥20 | 384 (86.3) | 252 (82.6) | 132 (94.3) | – |
| NA | 16 (3.6) | 16 (5.2) | 0 (0.0) | – |
| Lehmann subtype | – | – | – | 0.008 |
| BL1 | 58 (13.0) | 40 (13.1) | 18 (12.9) | – |
| BL2 | 19 (4.3) | 12 (3.9) | 7 (5.0) | – |
| IM | 68 (15.3) | 34 (11.1) | 34 (24.3) | – |
| LAR | 58 (13.0) | 37 (12.1) | 21 (15.0) | – |
| M | 51 (11.5) | 39 (12.8) | 12 (8.6) | – |
| MSL | 25 (5.6) | 16 (5.2) | 9 (6.4) | – |
| UNS | 42 (9.4) | 30 (9.8) | 12 (8.6) | – |
| NA | 124 (27.9) | 97 (31.8) | 27 (19.3) | – |
| FUSCCTNBC subtype | – | – | – | 0.019 |
| BLIS | 134 (30.1) | 95 (31.1) | 39 (27.9) | – |
| IM | 83 (18.7) | 48 (15.7) | 35 (25.0) | – |
| LAR | 79 (17.8) | 50 (16.4) | 29 (20.7) | – |
| MES | 50 (11.2) | 33 (10.8) | 17 (12.1) | – |
| NA | 99 (22.2) | 79 (25.9) | 20 (14.3) | – |
| RFS status at last follow-up | – | – | – | 0.286 |
| Event-free | 373 (83.8) | 260 (85.2) | 113 (80.7) | – |
| Event | 72 (16.2) | 45 (14.8) | 27 (19.3) | – |
| Family history | – | – | – | 0.181 |
| BC or OC | 49 (11.0) | 33 (10.8) | 16 (11.4) | – |
| Other cancer | 95 (21.3) | 58 (19.0) | 37 (26.4) | – |
| No | 301 (67.6) | 214 (70.2) | 87 (62.1) | – |
*, Student’s t-test [after Shapiro-Wilk test and analysis of variance (ANOVA)] was used to calculate P values. The other P values were calculated utilizing Pearson’s chi-square test or Fisher’s exact test. NA was not included in any calculation. IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; SD, standard deviation; FUSCCTNBC, Fudan University Shanghai Cancer Center Triple-negative Breast Cancer; RFS, relapse free survival; BC, breast cancer; OC, ovarian cancer.
Different molecular characteristics between LN positive and LN negative samples
We compared the mutational events in LN positive cases with those in LN negative cases. We found that mutations in TP53 and PIK3CA were the two most frequent in both LN positive and LN negative cases (Figure 2A). It is worth mentioning that WDR63, COL5A1, ATG2B, C17orf104, DDX41, F5 and LOXHD1 showed higher mutation frequencies in LN positive cases (Figure 2B). Most of them have been found playing important roles in the process of cancer metastasis, such as COL5A1 helping promoting gastric cancer metastasis (17) and the mutation of C17orf104 being found in the metastatic and recurrent head and neck squamous cell carcinoma (18). In addition, there were slightly more mutation events in LN positive cases (median =54) than that in LN negative cases (median =49), although there was no significant difference between them (Wilcoxon test, P>0.05) (Figure S1A,S1B).
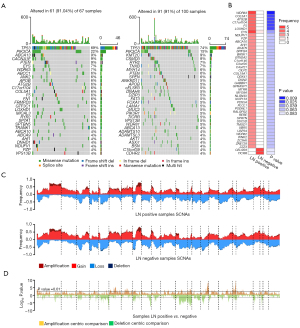
Next, we compared the SCNAs in LN positive cases with those in LN negative cases. We found that the frequencies of both amplification (68.9%) and deletion (82.3%) in SCNAs were higher in LN negative cases (Figure 2C,2D).
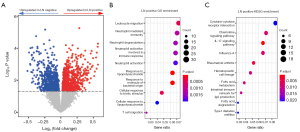
Finally, by analyzing the expression profile, genes (10.9%) were distinctly expressed between LN positive and LN negative cases (P<0.05). Among them, the expression levels of 1,954 genes were upregulated, and those of 1,466 genes were downregulated in LN positive cases (Figure 3A). To further investigate the features of these differentially expressed genes, we analyzed their enrichment pathways utilizing GO and KEGG gene sets. The results of GO enrichment analysis demonstrated that genes highly expressed in LN positive cases were mainly enriched in immunity and neutrophil-related pathways (Figure 3B). It has been proved that neutrophils and some special T cells conspired to promote breast cancer metastasis (19). In addition, we found that genes highly expressed in LN positive cases were enriched in cytokine-related pathways using KEGG enrichment analysis (Figure 3C).
Prediction models of axillary LNM developed by each omics
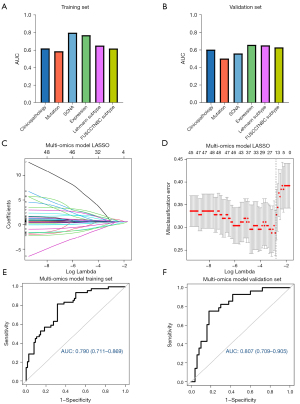
We tried to construct LNM prediction models based on the specific characteristics of each omics approach. All models were developed in the training set and were tested in the validation set. ROC curve analyses were utilized to evaluate the performance of each model (Figure 4A,4B).
Clinical model
Various clinical and pathological characteristics, including age at surgery, tumor size, histological grade, and HER2 and Ki-67 levels, were included to build a clinical model based on multivariable logistic regression analysis. The AUC was 0.624 (95% CI: 0.557–0.691) in the training set and 0.602 (95% CI: 0.502–0.702) in the validation set (Figure S2A,S2B).
Mutation model
We first selected 116 genes with a high frequency of mutation events (the number of mutations in each gene was greater than or equal to 3 in our cohort). We then used the LASSO regression model to select genes and established a panel of 12 genes (Figure S3A,S3B). Based on these results, we performed a multivariable logistic regression model with an area under the curve (AUC) of 0.591 (95% CI: 0.547–0.634) in the training set and 0.501 (95% CI: 0.444–0.558) in the validation set (Figure S3C,S3D).
SCNA model
To identify genes with differences in SCNA between LN positive and LN negative cases, Fisher’s exact test was utilized, and 1,008 genes (P<0.01) were selected. A LASSO regression model was used to further screen significant genes (Figure S4A,S4B), and 78 genes were finally selected to construct a multivariable logistic regression model. The AUC of the SCNA model was 0.805 (95% CI: 0.753–0.857) in the training set and only 0.558 (95% CI: 0.451–0.664) in the validation set (Figure S4C,S4D).
Expression model
At the transcriptomic level, we first selected 11 genes meeting two requirements: first, the expression was significantly distinct between LN positive and LN negative cases [|log2(fold change)| >0.3 and P<0.05]; second, the expression of genes in tumor tissues was different from that in normal tissues [|log2(fold change)| >2 & P<0.01]. Then, we performed a LASSO regression model, after which 10 genes (Figure S5A,S5B) were selected for the multivariable logistic regression model. The AUC of the model was 0.777 (95% CI: 0.718–0.837) in the training set and 0.656 (95% CI: 0.557–0.755) in the validation set (Figure S5C,S5D).
Molecular subtype model
In Lehmann’s research, TNBC could be divided into 7 subtypes (BL1, BL2, IM, LAR, M, MSL and UNS) according to RNA expression (Table 1) (20). Based on the Lehmann subtype, we established a model to predict LN status. The AUC of the model was 0.656 (95% CI: 0.582–0.730) in the training set and 0.650 (95% CI: 0.548–0.752) in the validation set (Figure S6A,S6B).
In our previous study, through RNA-sequencing, TNBC was also divided into 4 subtypes (BLIS, IM, LAR and MES) (12). We then built a model based on FUSCCTNBC subtypes, with an AUC of 0.623 (95% CI: 0.549–0.697) in the training set and 0.627 (95% CI: 0.527–0.726) in the validation set (Figure S6C,S6D).
Construction and performance of the multi-omics prediction model
To obtain a model with a better predictive performance, we combined all predictive markers identified above, including 5 clinicopathologic characteristics, 12 gene mutations, 78 SCNA features, the RNA expression of 10 selected genes and two kinds of TNBC subtypes. From all 107 predictive markers, we built a LASSO regression model to establish a signature with a panel of 17 features (Figure 4C,4D). To further screen these features, forward-stepwise selection was employed by utilizing IBM SPSS Statistics (R23.0.0.0), and 5 predictive markers, including tumor size, SCNAs of ZBTB6 and MTHFD1, and mRNA expression levels of GLP1R and NPY5R, were finally confirmed and used to construct the multi-omics prediction model. Univariate logistic regression was used to evaluate the risk of the 5 markers (Table 2). Then, we performed multivariable logistic regression to build an integrated multi-omics model.
Table 2
| Variable | Training set (n=318) | Validation set (n=145) | |||
|---|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | ||
| Tumor size (cm) | 1.38 (0.98–1.95) | 0.064 | 3.27 (1.62–6.61) | 0.001 | |
| ZBTB6 (copy number) | 4.9 (1.41–16.95) | 0.012 | 2.67 (0.69–10.23) | 0.153 | |
| MTHFD1 (copy number) | 5.6 (1.86–16.85) | 0.002 | 2.42 (0.81–7.27) | 0.114 | |
| GLP1R (expression) | 0.15 (0.04–0.5) | 0.002 | 0.11 (0.02–0.74) | 0.023 | |
| NPY5R (expression) | 0.27 (0.12–0.6) | 0.001 | 0.5 (0.21–1.2) | 0.121 | |
The AUC of the multi-omics prediction model was 0.790 (95% CI: 0.711–0.869) in the training set and 0.807 (95% CI: 0.709–0.905) in the validation set (Figure 4E,4F). The corresponding calibration curves were plotted to assess the calibration of the multi-omics model in both the training and validation sets, and the Hosmer-Lemeshow test indicated that there was no significant departure from excellent fit (P>0.05) (Figure S7A,S7B). In addition, the decision curve analysis (DCA) results for the multi-omics model were also plotted to show its net benefit (Figure S7C,S7D).
Finally, in the fully integrated approach, we took all multi-omics factors into the integrated model to apply integration methods. Through LASSO regression (Figure S8A,S8B) and forward-stepwise selection, potential predictors were selected, and a logistic regression model was built. The model had a good performance as well (AUC of 0.83 in the training set, AUC of 0.73 in the validation set) (Figure S8C,S8D), which further proved that predicting LNM using multi-omics was practicable.
Discussion
Currently, according to preoperative evaluation by various methods, some breast cancer patients are exempt from SLNB. This reduces the pain resulting from invasive procedures, while retaining the risk of underestimation of axillary LNM. Therefore, more tools are needed to accurately assess the risk of ALN metastasis and select the proper patient eligible for SLNB exemption. The LN status of TNBC is difficult to predict using clinical factors only. We aim to provide more evidence for TNBC patients regarding whether they could be exempted from LN surgical excision. In this study, we included clinicopathological information and genomic and transcriptomic data. Through analysis and comparison, we found that there were distinctive characteristics between LN positive and LN negative cases in the four omics datasets. We then selected the most pivotal characteristics of each omics dataset further to construct an integrated multi-omics model, which performed better than single omics-based models.
Moreover, we found that different omics techniques showed distinct impacts on LNM. The predictive power of the clinical model was not satisfactory in the training set or validation set. In contrast to other subtypes of breast cancer, TNBC is a group of diseases with significant inter-tumor heterogeneity (12) and a high degree of malignancy (21), and its LNM is hard to predict using clinical factors only. Compared with LN negative cases, LN positive cases had more mutational events. However, among all models in the study, the performance of the mutation-based model was the worst, and its AUC was low in neither the training set nor the validation set. This might be caused by the low mutation frequency of each gene. The majority of the genes were mutated in less than 5% of TNBCs, while a large fraction of them might be “passenger” events without specific biological impact (22). Taken together, somatic mutations in TNBC cannot reflect the differences well between LN positive and LN negative cases. In our previous studies, we found that TNBC is a kind of cancer significantly driven by SCNA (12,16,23). Therefore, we analyzed the SCNA of each gene in the study cohort, and we found that there was a great difference between LN positive and LN negative cases. In the SCNA-based model, the performance was excellent in the training set but mediocre in the validation set, which may be due to the overfitting of the model with an excessive number of included genes. Among the four omics approaches, transcriptomic data showed the most significant differences. There were 1954 upregulated genes and 1,466 downregulated genes in LN positive cases. At the same time, the transcriptomic model also showed the best predictive performance, the AUC of which was 0.777 in the training set and 0.656 in the validation set, indicating that the transcriptome could better distinguish the internal differences between LN positive and LN negative cases. Then, we found that it was also difficult to predict LN status using only the information of TNBC Lehmann or FUSCC subtypes.
In this study, a total of 5 predictive factors were finally included for the construction of the multi-omics model, including tumor size, SCNA of ZBTB6 and MTHFD1, and the RNA expression levels of GLP1R and NPY5R. Among them, tumor size and SCNA of ZBTB6 and MTHFD1 are positively correlated with the risk of LNM. Both tumor size and lymph node status are important factors for evaluating the tumor malignancy. Many studies have already demonstrated that tumor sizes are directly related to LNM in breast cancer, especially in hormone receptor-positive breast cancers (10,24-26). In our research, we found that tumor size was associated with LNM in TNBC. However, it was demonstrated that tumor size along with other clinicopathological factors were not sufficient to predict LNM, and the AUC of its model was just 0.624 in the training set and 0.602 in the validation set. Hence, we hoped to seek more predictive factors in more dimensions. In this study, we found that the amplification of ZBTB6 was related to a high risk of LNM. There have been few studies of ZBTB6, a gene related to energy metabolism, and it has only been reported to be a prognostic indicator in esophageal cancer (27). Here, we found that the amplification of ZBTB6 was related to a high risk of LNM in TNBC, but the mechanism is still unknown. MTHFD1, as a well-known gene related to folic acid metabolism, has been found to interact with BRD4, participate in folic acid metabolism and transcriptional regulation (28), and play an important role in many cancers, such as melanoma, lung cancer and colorectal cancer (28-35). MTHFD1 was also found to promote the progression of breast cancer (29). Consistent with our study, the amplification of MTHFD1 was related to a high risk of LNM in TNBC.
Among the 5 predictors, the RNA expression levels of GLP1R and NPY5R were negatively correlated with the risk of LNM. GLP1R is also a well-studied and powerful gene that has been found to activate cAMP and inhibit the proliferation of breast cancer (36). In our multi-omics model, the expression of GLP1R negatively contributed to the LNM risk score. There have been few studies on NPY5R, and no related mechanism has been studied in the tumor field. Similar to GLP1R, the high expression of NPY5R was a protective factor against LNM in the model. Overall, these molecules, which we selected as the most significant predictive markers of axillary LNM, are still lacking in mechanistic explorations of tumors and need further study.
Although the multi-omics model has robust predictive efficacy and is feasible in theory, it is difficult to implement in clinical practice. Neither genome nor transcriptome sequencing data can be obtained in a short time, and the financial cost is high. However, with the development of sequencing technology, it remains to be seen whether sequencing technology can achieve economic timeliness in the future. Based on this model, we also plan to evaluate the level of these genes through rapid methods, such as quantitative polymerase chain reaction (qPCR) or immunohistochemistry (IHC).
Our study had several limitations. First, the models were hard to validate externally because of the lack of full clinicopathological information in public datasets and the batch effect of sequencing platforms. To note, we plan to validate the multi-omics model through a prospective study in the future. Second, although it is the largest TNBC multi-omics cohort analyzed in the study, the number of patients is limited, and the reliability of the model needs to be further improved by increasing the number of cases. Finally, the biological impact of the selected predictors needs further investigation.
Despite the limitations, to our knowledge, this is the first study to predict LN status in TNBC utilizing an integrated multi-omics model, which performed better than models based on each single omics.
Conclusions
To conclude, we compared the difference between LN positive and LN negative patients using the largest multi-omics TNBC cohort and identified clinicopathological, genomic and transcriptomic characteristics potentially related to LNM. Based on these indicators, we established an integrated multi-omics model with robust performance, showing an AUC of 0.796 in the training set and 0.807 in the validation set, which proved that using multi-omics data could better predict LNM in TNBC. Importantly, this will help us achieve a more precise management of lymph node metastasis in TNBC patients.
Acknowledgments
Funding: This work was supported by grants from the National Key Research and Development Project of China (No. 2020YFA0112304), the National Natural Science Foundation of China (Nos. 81874113, 82002802, 91959207), the Shanghai Key Laboratory of Breast Cancer (No. ZDSYS2101), the Shanghai Key Clinical Specialty of Oncology (No. shslczdzk02001), the Clinical Research Plan of SHDC (No. SHDC2020CR5005) and Shanghai Sailing Program for Youth S&T Talents (No. 20YF1408700). The funders had no role in the design of the study and collection, analysis, interpretation of data or writing the manuscript.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-277/rc
Peer Review File: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-277/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-277/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Clark GM. Prognostic and Predictive Factors for Breast Cancer. Breast Cancer 1995;2:79-89. [Crossref] [PubMed]
- Galimberti V, Cole BF, Zurrida S, et al. Axillary dissection versus no axillary dissection in patients with sentinel-node micrometastases (IBCSG 23–01): a phase 3 randomised controlled trial. Lancet Oncol 2013;14:297-305. [Crossref] [PubMed]
- Giuliano AE, Hunt KK, Ballman KV, et al. Axillary dissection vs no axillary dissection in women with invasive breast cancer and sentinel node metastasis: a randomized clinical trial. JAMA 2011;305:569-75. [Crossref] [PubMed]
- Cawley J, Willage B, Frisvold D. Pass-Through of a Tax on Sugar-Sweetened Beverages at the Philadelphia International Airport. JAMA 2018;319:305-6. [Crossref] [PubMed]
- Giuliano AE, Ballman KV, McCall L, et al. Effect of Axillary Dissection vs No Axillary Dissection on 10-Year Overall Survival Among Women With Invasive Breast Cancer and Sentinel Node Metastasis: The ACOSOG Z0011 (Alliance) Randomized Clinical Trial. JAMA 2017;318:918-26. [Crossref] [PubMed]
- Tadros AB, Yang WT, Krishnamurthy S, et al. Identification of Patients With Documented Pathologic Complete Response in the Breast After Neoadjuvant Chemotherapy for Omission of Axillary Surgery. JAMA Surg 2017;152:665-70. [Crossref] [PubMed]
- Latosinsky S, Dabbs K, Moffat F, et al. Canadian Association of General Surgeons and American College of Surgeons Evidence-Based Reviews in Surgery. 27. Quality-of-life outcomes with sentinel node biopsy versus standard axillary treatment in patients with operable breast cancer. Randomized multicenter trial of sentinel node biopsy versus standard axillary treatment in operable breast cancer: the ALMANAC Trial. Can J Surg 2008;51:483-5. [PubMed]
- Boone BA, Huynh C, Spangler ML, et al. Axillary Lymph Node Burden in Invasive Breast Cancer: A Comparison of the Predictive Value of Ultrasound-Guided Needle Biopsy and Sentinel Lymph Node Biopsy. Clin Breast Cancer 2015;15:e243-8. [Crossref] [PubMed]
- Cui X, Zhu H, Huang J. Nomogram for Predicting Lymph Node Involvement in Triple-Negative Breast Cancer. Front Oncol 2020;10:608334. [Crossref] [PubMed]
- Dihge L, Vallon-Christersson J, Hegardt C, et al. Prediction of Lymph Node Metastasis in Breast Cancer by Gene Expression and Clinicopathological Models: Development and Validation within a Population-Based Cohort. Clin Cancer Res 2019;25:6368-81. [Crossref] [PubMed]
- Tan W, Xie X, Huang Z, et al. Construction of an immune-related genes nomogram for the preoperative prediction of axillary lymph node metastasis in triple-negative breast cancer. Artif Cells Nanomed Biotechnol 2020;48:288-97. [Crossref] [PubMed]
- Jiang YZ, Ma D, Suo C, et al. Genomic and Transcriptomic Landscape of Triple-Negative Breast Cancers: Subtypes and Treatment Strategies. Cancer Cell 2019;35:428-40.e5. [Crossref] [PubMed]
- Gene Ontology C. The Gene Ontology project in 2008. Nucleic Acids Res 2008;36:D440-4. [Crossref] [PubMed]
- Okuda S, Yamada T, Hamajima M, et al. KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res 2008;36:W423-6. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Xiao Y, Ma D, Zhao S, et al. Multi-Omics Profiling Reveals Distinct Microenvironment Characterization and Suggests Immune Escape Mechanisms of Triple-Negative Breast Cancer. Clin Cancer Res 2019;25:5002-14. [Crossref] [PubMed]
- Zhang Y, Jing Y, Wang Y, et al. NAT10 promotes gastric cancer metastasis via N4-acetylated COL5A1. Signal Transduct Target Ther 2021;6:173. [Crossref] [PubMed]
- Hedberg ML, Goh G, Chiosea SI, et al. Genetic landscape of metastatic and recurrent head and neck squamous cell carcinoma. Journal of Clinical Investigation 2015;126:169-80. [Crossref] [PubMed]
- Coffelt SB, Kersten K, Doornebal CW, et al. IL-17-producing gammadelta T cells and neutrophils conspire to promote breast cancer metastasis. Nature 2015;522:345-8. [Crossref] [PubMed]
- Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011;121:2750-67. [Crossref] [PubMed]
- Carey L, Winer E, Viale G, et al. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol 2010;7:683-92. [Crossref] [PubMed]
- Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. [Crossref] [PubMed]
- Ma D, Chen SY, Ren JX, et al. Molecular Features and Functional Implications of Germline Variants in Triple-Negative Breast Cancer. J Natl Cancer Inst 2021;113:884-92. [Crossref] [PubMed]
- Bundred NJ. Prognostic and predictive factors in breast cancer. Cancer Treat Rev 2001;27:137-42. [Crossref] [PubMed]
- Cianfrocca M, Goldstein LJ. Prognostic and predictive factors in early-stage breast cancer. Oncologist 2004;9:606-16. [Crossref] [PubMed]
- Martin M, Gonzalez Palacios F, Cortes J, et al. Prognostic and predictive factors and genetic analysis of early breast cancer. Clin Transl Oncol 2009;11:634-42. [Crossref] [PubMed]
- Zheng W, Chen C, Yu J, et al. An energy metabolism-based eight-gene signature correlates with the clinical outcome of esophagus carcinoma. BMC Cancer 2021;21:345. [Crossref] [PubMed]
- Sdelci S, Rendeiro AF, Rathert P, et al. MTHFD1 interaction with BRD4 links folate metabolism to transcriptional regulation. Nat Genet 2019;51:990-8. [Crossref] [PubMed]
- Cao S, Zhu Z, Zhou J, et al. Associations of one-carbon metabolism-related gene polymorphisms with breast cancer risk are modulated by diet, being higher when adherence to the Mediterranean dietary pattern is low. Breast Cancer Res Treat 2021;187:793-804. [Crossref] [PubMed]
- Chen K, Wu S, Ye S, et al. Dimethyl Fumarate Induces Metabolic Crisie to Suppress Pancreatic Carcinoma. Front Pharmacol 2021;12:617714. [Crossref] [PubMed]
- Collin SM, Metcalfe C, Zuccolo L, et al. Association of folate-pathway gene polymorphisms with the risk of prostate cancer: a population-based nested case-control study, systematic review, and meta-analysis. Cancer Epidemiol Biomarkers Prev 2009;18:2528-39. [Crossref] [PubMed]
- Levesque N, Christensen KE, Van Der Kraak L, et al. Murine MTHFD1-synthetase deficiency, a model for the human MTHFD1 R653Q polymorphism, decreases growth of colorectal tumors. Mol Carcinog 2017;56:1030-40. [Crossref] [PubMed]
- Moussa C, Ross N, Jolette P, et al. Altered folate metabolism modifies cell proliferation and progesterone secretion in human placental choriocarcinoma JEG-3 cells. Br J Nutr 2015;114:844-52. [Crossref] [PubMed]
- Yao S, Peng L, Elakad O, et al. One carbon metabolism in human lung cancer. Transl Lung Cancer Res 2021;10:2523-38. [Crossref] [PubMed]
- Piskounova E, Agathocleous M, Murphy MM, et al. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature 2015;527:186-91. [Crossref] [PubMed]
- Ligumsky H, Wolf I, Israeli S, et al. The peptide-hormone glucagon-like peptide-1 activates cAMP and inhibits growth of breast cancer cells. Breast Cancer Res Treat 2012;132:449-61. [Crossref] [PubMed]


