Toward precision medicine in neurological diseases
Introduction
With the Human Genome Project going successfully, the concept that treatments of individuals could be defined individually has often been espoused but rarely realized. The initiation of next-generation sequencing (NGS) has paved a new way of this concept, and a new name to go with it-precision medicine. Precision medicine refers to the scientific basis that underpins the personalization of medical care, particularly in the context of treatments targeted towards the precise molecular causes of disease (1). Its core, in which prevention and treatment strategies are based on taking individual variability into account, has been applied in clinical practice for centuries. Unlike personalized medicine, precision medicine emphasizes an innovative approach based on specific characteristics including genetic, biomarker and psychosocial of each patient. The realization of precision medicine is perhaps best illustrated in cancer which is multigenetic, and its mechanism-based treatments have successfully moved from bench to bedside (2). Likewise, in most therapeutic areas, particularly the clinically multigenic neurodegenerative disease with a strong genetic component in neurological diseases, precision medicine remains aspirational.
Here we propose that, after cancer, neurological diseases also offer the most compelling opportunities to achieve precision medicine for the following reasons: the rapid progress in the gene discovery; the ability to assess targeted diagnostic biomarkers in large-scale biologic databases or clinical trials and the initiation of NGS allowing the development of targeted pathways tailored to genetically defined treatment of the neurological diseases. To realize the potential of precision medicine in neurological diseases, however, many distinct areas of basic and translational research must collaborate and cooperate together. Here, we outline a strategy for the progress of an integrated program for precision medicine in neurological diseases, including the genome-wide association studies (GWAS), the development of diagnostic biomarkers in large-scale biologic databases or clinical trials, and potential treatment of neurological diseases. We conclude that fostering integrated research teams to advance precision medicine in neurological diseases will improve health care and that neurological diseases could serve as models for other therapeutic areas (Figure 1).
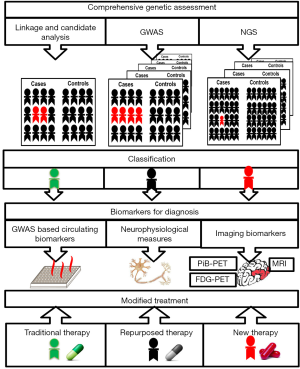
Precision genetics for precision medicine
Neurological diseases are ideal choices for enhancing the impact of precision medicine. Precision medicine has recently become popular especially among those scientists and clinicians who are applying genomics and data mining to classify individuals into subgroups with different susceptibility to neurological diseases. Many neurodegenerative diseases, such as Alzheimer’s disease (AD), Parkinson’s disease (PD), Amyotrophic lateral sclerosis (ALS), are clinically heterogeneous neurodegenerative disease with a strong genetic component (3-7). Besides, gene polymorphism also plays an irreplaceable role in stroke and epilepsy (8). These are the end results of the complex effects of several genes interacting with the environment (9). In view of their high population prevalence, to identify its genetic background is as important as finding the environmental factors. Living in the era of genomics and big data, varied genetic research of neurological diseases will propel our understanding—their origins and mechanisms, and opportunities for prevention and treatment—laying a firm, broad foundation for precision medicine. Although the investigation of clinically relevant genetic variation remains a challenge, established examples are being used to stratify subgroups of patients to identify optimal treatments (10).
Emerging new tools, such as microarray and NGS, make it possible to collect large amounts of digital genetic data. These new sources of data, together with increasingly available molecular information from the genome, transcriptome, epigenome and proteome, have created much value and interest in treatments of neurological diseases. Before the development of NGS, both linkage analyses and targeted candidate gene studies identified a number of neurological diseases candidate genes. One of the earliest ways into understanding the genetic basis of neurological diseases was through investigating the association between neurological diseases and a specific allele of a single nucleotide polymorphism within the functional candidate genes between patients and controls. For example, much of the initial work on candidate genes of AD has come from studies of rare families where early-onset AD is transmitted autosomal-dominantly, resulting from three genes: APP, PSEN1 and PSEN2. These mutations alter production of the amyloid-β (Aβ) peptide which is the principal component of senile plaques (11). However, genetic variation in the APP, PSEN1 and PSEN2 genes has little or no effect on late-onset AD (12). In the search for new genes in AD, classic linkage-based and candidate-gene-based association studies have been supplanted by exome sequencing studies and GWAS. The hypothesis-free approach by GWAS contains nearly all common variants in the entire genome which can be tested for association with AD (13). GWAS allows interrogation of millions of polymorphisms simultaneously across the genome, leading to renewed hope, especially the era of large-scale GWAS. Recent large-scale GWAS have identified millions of genetic variations simultaneously across the genome and have been the driving force in the identification of more than ten main risk genes (CLU, TREM2, CR1, PICALM, BIN1, MS4A, CD2AP, CD33, EPHA1 and ABCA7) for late-onset AD (9,12-17). Tremendous progress has been made in understanding genomics of PD since the identification of the mutation in the gene encoding α-synuclein as well. Like AD, large consortia have pursued genetic variants associated with PD using genomic approaches and have identified a similar number of risk loci (18,19).
Encouragingly, rapid improvements in sequencing have driven genetic discovery in neurological diseases. The genome, transcriptome and the epigenome are three major categories of information that can be acquired through various GWAS and NGS-based assays. Although the number of neurological diseases genes is too large for sequencing for diagnostic purposes, NGS techniques enable simultaneous screening of a whole battery of genes implicated in neurological diseases. In addition to detecting novel risk factors in large samples, NGS approaches can deliver novel insights with even small numbers of patients. Furthermore, understanding the mechanisms how neurons alter and maintain their molecular signatures during disease processing is a fundamental goal of neurological diseases. Several genes, including ABCA7 (20), BIN1 (21), CR1 (22), PLD3 (23) and PICALM (24), were revealed as the excess burden of deleterious coding mutations in AD. In the epileptic encephalopathies alone, trio sequencing has led to the identification of ALG, GABRB3, DNM1, HCN1, GRIN2A, GABRA1, GNAO1, KCNT1, SCN2A, SCN8A, and SLC35A2 as genes associated with epilepsy while many of the proteins encoded by these genes are involved in synaptic transmission (8). The identification of these rare genes has opened new avenues for exploration of the underlying disease mechanisms. Besides, NGS technology is rapidly transforming the ability to probe the molecular basis of neuronal function. RNA-sequencing (RNA-seq) can supply information regarding transcript structure, including single base-pair resolution of transcript boundaries and boundaries between exons. Differential transcription is examined using RNA-seq in AD brain tissue, which allows for the examination of protein coding genes, non-coding RNAs, and splicing. Networks will be discovered and this may contribute to neurological diseases mechanisms. Likewise, in animal models or blood samples, noncoding RNAs were also observed participating in AD progression (25). NGS can define not only the complete molecular signatures of cells by transcriptome analyses, but also the cascade of events that induce or maintain such signatures by epigenetic analyses. Epigenetics refers to changes in gene regulation brought about through modifications to the DNA’s packaging proteins or the DNA molecules themselves without changing the underlying sequence (26). Such changes are thought to be a way in which the environment can influence genetics, bringing about potentially pathogenic alterations in the way genes are expressed. These studies provide compelling evidence for genuine, genome-wide significant associations between differentially methylated regions and neurological diseases, and suggest that the correlation relationship could reflect a causative one. Collectively, these findings could lead researchers to focus on rare variants in neurological diseases precisely when developments in NGS facilitated the comprehensive interrogation of genomes. In a word, GWAS and NGS will greatly help with personalization of treatment for neurological diseases, particularly when treatments that target the underlying molecular causes rather than the clinical symptoms become available.
Classification, biomarkers and modified treatment modalities
Ideally, precision medicine contrasts with the traditional medicine in phenotypic classification based on genetic risk, and intervention to suppress pathophysiologic processes while still latent (27). To date, all medical disciplines are seeking to develop biomarkers to detect latent pathophysiologic processes in the expectation that the earliest detection provides the greatest opportunity for effective intervention. Due to the sources of big data analyzed by computational tools and newly available genetic technologies, targeted treatments are available for genetically defined subgroups of patients, enabling the provision of the right medication at the right dose for the right patient (28). In such a circumstance, patients are not only categorized by traditional subgroups, but also treated as individual cases based on the multi-scale data. The data will be analyzed according to genomics, transcriptome, as well as other molecular and biochemical system. The computational approaches incorporate a wide range of these personalized data sources to establish the patient’s data at the molecular levels, covering traditional medical information (the history, physical examination, and laboratory tests), imaging, genomic and other analyses, to individualized objective phenotypic data on function and overall health status by mobile health technology (29). It will expand the scope of precision medicine and deepen our insight into it by redefining the classification of neurological diseases, interpreting these big data with important prognostic and treatment implications, and facilitating their application in the clinical setting.
Biomarkers that have prognostic value for survival would be of value for decision-making and planning of care. Technological developments have led to the discovery of many agents in biofluids neurophysiological measures and neuroimaging biomarkers for neurological diseases. There is an urgent need to find and validate biomarkers to predict future clinical decline. Investigators from academia and the pharmaceutical industry became interested in how “disease modification” of neurological diseases could be detected using a variety of biomarkers, including volumetric magnetic resonance imaging (MRI), fluorodeoxyglucose positron emission tomography (PET), blood and cerebrospinal fluid analyses (25,30-34). Besides, motor unit number index, which enables quantification and tracing of motor unit numbers, might differentiate ALS from other diseases and enable disease progression to be monitored (35). What’s more, a combination of viscoelastic methodologies including thromboelastography and scanning electron microscopy approach was validated useful to stroke patients in a personalized patient-centered regime (34).
Neuroimaging offers a noninvasive approach to biomarker discovery and disease monitoring not only in stroke (36-38) but also in other neurodegenerative disorders. The dopamine terminal dysfunction can be demonstrated by PET or single photoemission computed tomography with different tracers, which contribute to early and accurate diagnosis leading to appropriate medications in PD. Voxel and surface-based MRI quantification demonstrated thinning of the primary motor cortex as well. However, the correlation of imaging metrics with the absolute level of disability of MRI, using estimated rate of disease progression, was inconsistent in ALS (39). Intriguingly, these neuroimaging methods are likely to be shaped by genetic influences with heritability (32). Improved neuroimaging of AD have aided diagnosis of AD in the very early stages, and have facilitated differential diagnosis between AD and other neurodegenerative disorders with dementia (32). Evaluation of specific molecules by biochemical assay has been comparably successful in the research area (25), and both are being considered for more widespread application (40). Quantification of molecules singlely or in large groups in other biofluids, such as serum, plasma, has been investigated many times, although none has yet revealed an appropriate biomarker.
After comprehensive genetic classification and accurate biomarker for latent pathophysiologic processes, the promise of precision medicine culminates in treatments that prevent, stop, or slow progression based on an individual’s specialty. However, there are no widely-accepted cured therapies for most neurological diseases. New candidate treatments for genetic subtypes of neurological diseases are in urgent need. Thus re-tasking the technical capacity and expertise from the neurological diseases genome atlas to expand neurological diseases genomic sequencing efforts yet further to allow a big-data-based approach that links knowledge of variation and acquired somatic mutations with clinical phenotype is intellectually appealing. Furthermore, in terms of drug application, patients with the same or similar symptoms may use the same treatment. In fact, different patients have different sensitivity to the same drug. The differences between individuals are closely related to their different genetic background. Precise medical treatment can be based on molecular characteristics of patients with a detailed classification and provide a reference for effective treatment. As neurological diseases require multiple targets for drug therapy, integrated analysis of studies and clinical data to identify targets is needed. To facilitate the development of new treatments for genetic neurological diseases, a clear functional understanding of the mutation that is both related to how the mutation causes disorders and whether it is amenable to screen is essential. AD is mainly characterized by amyloid deposition and tau phosphorylation, so the development of anti-AD drugs should mainly be used to reduce amyloid deposition. Many disease-modifying strategies targeting Aβ peptide and abnormal tau protein, as well as providing neuroprotective effects are currently under active evaluation (41). Examples of effective precision therapy in genetic epilepsy are already emerging including the mutations in SLC2A1 and ALDH7A1 (42). Subsequent multiple clinical trials to test the proposed disease-altering interventions have been attempted but ultimately failed, as they neglected to consider the underlying clinical and biological complexity of the disease. With precision medicine has been overlooked recently, clinical trials are emerging as disease related genes are discovered continuously, such as SOD1, C9orf72, TDP-43 and ATXN2 targeted therapeutic strategies in ALS (5). In addition, adaptive deep brain stimulation can provide a precision and individualized treatment for PD patient through automatically adjusting parameters according to the real-time brain/body response.
Conclusions
Precision medicine is emerging as a natural product that integrates basic research and clinical practice to build a platform that can better guide individualized patient care. The advantage for the concept of “precision medicine” is the emerging approaches and technologies which enabled tailored treatments targeted to the needs of individuals according to the precise genetic, biomarker characteristics and bioinformatics that underpin the personalization of medical care. The considerations and examples outlined above make it clear that precision medicine could transform clinical care in neurological diseases, and could lead to a new treatment framework for the neurological diseases. We propose four crucial elements of precision medicine for neurological diseases: comprehensive genetic assessment, classification, biomarkers, and treatments specified to a person’s molecular drivers. From wearable activity trackers to meta-genomic sequencing, we are able to monitor our personal environment and health more than before. Potential toxicities of the treatment and the severity and prevalence of the disorder must be considered in the search for candidate treatments for genetic subtypes of neurological diseases. The prospect of applying this concept broadly has been dramatically improved by the recent development of large-scale biologic databases (such as the human genome sequence), powerful methods for characterizing patients (such as proteomics, metabolomics, genomics, diverse cellular assays, and even mobile health technology), and computational tools for analyzing large sets of data. However, encouragement of creative approaches to precision medicine and the ultimate usage still needs to guide clinical practice.
Acknowledgements
Funding: This work was supported by grants from the National Natural Science Foundation of China (81000544, 81171209, 81571245, 81501092 and 81501103), the Shandong Provincial Outstanding Medical Academic Professional Program, Qingdao Key Health Discipline Development Fund, Qingdao Outstanding Health Professional Development Fund, and Shandong Provincial Collaborative Innovation Center for Neurodegenerative Disorders.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Moving toward precision medicine. Lancet 2011;378:1678. [Crossref] [PubMed]
- Collisson EA, Cho RJ, Gray JW. What are we learning from the cancer genome? Nat Rev Clin Oncol 2012;9:621-30. [Crossref] [PubMed]
- Gatz M, Reynolds CA, Fratiglioni L, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry 2006;63:168-74. [Crossref] [PubMed]
- Montine TJ, Montine KS. Precision medicine: clarity for the clinical and biological complexity of Alzheimer's and Parkinson's diseases. J Exp Med 2015;212:601-5. [Crossref] [PubMed]
- Zou ZY, Liu CY, Che CH, et al. Toward precision medicine in amyotrophic lateral sclerosis. Ann Transl Med 2016;4:27. [PubMed]
- Bu LL, Yang K, Xiong WX, et al. Toward precision medicine in Parkinson's disease. Ann Transl Med 2016;4:26. [PubMed]
- Jiang T, Yu JT, Tian Y, et al. Epidemiology and etiology of Alzheimer's disease: from genetic to non-genetic factors. Curr Alzheimer Res 2013;10:852-67. [Crossref] [PubMed]
- Epi PM. Consortium. A roadmap for precision medicine in the epilepsies. Lancet Neurol 2015;14:1219-28. [Crossref] [PubMed]
- Seshadri S, Fitzpatrick AL, Ikram MA, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 2010;303:1832-40. [Crossref] [PubMed]
- Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 2015;372:793-5. [Crossref] [PubMed]
- Tanzi RE, Bertram L. Twenty years of the Alzheimer's disease amyloid hypothesis: a genetic perspective. Cell 2005;120:545-55. [Crossref] [PubMed]
- Harold D, Abraham R, Hollingworth P, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet 2009;41:1088-93. [Crossref] [PubMed]
- Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A 2009;106:9362-7. [Crossref] [PubMed]
- Hollingworth P, Harold D, Sims R, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet 2011;43:429-35. [Crossref] [PubMed]
- Jun G, Ibrahim-Verbaas CA, Vronskaya M, et al. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol Psychiatry 2016;21:108-117. [Crossref] [PubMed]
- Lambert JC, Heath S, Even G, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet 2009;41:1094-9. [PubMed]
- Naj AC, Jun G, Beecham GW, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet 2011;43:436-41. [Crossref] [PubMed]
- International Parkinson Disease Genomics Consortium, Nalls MA, Plagnol V, et al. Imputation of sequence variants for identification of genetic risks for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet 2011;377:641-9. [Crossref] [PubMed]
- Lill CM, Roehr JT, McQueen MB, et al. Comprehensive research synopsis and systematic meta-analyses in Parkinson's disease genetics: The PDGene database. PLoS Genet 2012;8:e1002548. [Crossref] [PubMed]
- Vardarajan BN, Ghani M, Kahn A, et al. Rare coding mutations identified by sequencing of Alzheimer disease genome-wide association studies loci. Ann Neurol 2015;78:487-98. [PubMed]
- Tan MS, Yu JT, Jiang T, et al. Genetic variation in BIN1 gene and Alzheimer's disease risk in Han Chinese individuals. Neurobiol Aging 2014;35:1781.e1-8.
- Ma XY, Yu JT, Tan MS, et al. Missense variants in CR1 are associated with increased risk of Alzheimer' disease in Han Chinese. Neurobiol Aging 2014;35:443.e17-21.
- Cacace R, Van den Bossche T, Engelborghs S, et al. Rare variants in PLD3 do not affect risk for early-onset Alzheimer disease in a European consortium cohort. Hum Mutat 2015;36:1226-35. [Crossref] [PubMed]
- Jiang T, Yu JT, Tan MS, et al. Genetic variation in PICALM and Alzheimer's disease risk in Han Chinese. Neurobiol Aging 2014;35:934.e1-3.
- Tan L, Yu JT, Tan MS, et al. Genome-wide serum microRNA expression profiling identifies serum biomarkers for Alzheimer's disease. J Alzheimers Dis 2014;40:1017-27. [PubMed]
- Mazzio EA, Soliman KF. Basic concepts of epigenetics: impact of environmental signals on gene expression. Epigenetics 2012;7:119-30. [Crossref] [PubMed]
- Cholerton B, Larson EB, Quinn JF, et al. Precision medicine: clarity for the complexity of dementia. Am J Pathol 2016;186:500-6. [Crossref] [PubMed]
- Mohler J, Najafi B, Fain M, et al. Precision medicine: a wider definition. J Am Geriatr Soc 2015;63:1971-2. [Crossref] [PubMed]
- Jameson JL, Longo DL. Precision medicine--personalized, problematic, and promising. N Engl J Med 2015;372:2229-34. [Crossref] [PubMed]
- Li JQ, Wang HF, Zhu XC, et al. GWAS-linked loci and neuroimaging measures in Alzheimer's disease. Mol Neurobiol 2016. [Epub ahead of print]. [PubMed]
- Steinacker P, Feneberg E, Weishaupt J, et al. Neurofilaments in the diagnosis of motoneuron diseases: a prospective study on 455 patients. J Neurol Neurosurg Psychiatry 2016;87:12-20. [PubMed]
- Weiner MW, Veitch DP, Aisen PS, et al. 2014 Update of the Alzheimer's Disease Neuroimaging Initiative: A review of papers published since its inception. Alzheimers Dement 2015;11:e1-120. [Crossref] [PubMed]
- Dubois B, Feldman HH, Jacova C, et al. Advancing research diagnostic criteria for Alzheimer's disease: the IWG-2 criteria. Lancet Neurol 2014;13:614-29. [PubMed]
- de Villiers S, Swanepoel A, Bester J, et al. Novel diagnostic and monitoring tools in stroke: an individualized patient-centered precision medicine approach. J Atheroscler Thromb 2015. [Epub ahead of print]. [Crossref] [PubMed]
- Neuwirth C, Barkhaus PE, Burkhardt C, et al. Tracking motor neuron loss in a set of six muscles in amyotrophic lateral sclerosis using the Motor Unit Number Index (MUNIX): a 15-month longitudinal multicentre trial. J Neurol Neurosurg Psychiatry 2015;86:1172-9. [Crossref] [PubMed]
- Feldmann E, Liebeskind DS. Developing precision stroke imaging. Front Neurol 2014;5:29. [Crossref] [PubMed]
- Liebeskind DS, Feldmann E. Imaging of cerebrovascular disorders: precision medicine and the collaterome. Ann N Y Acad Sci 2015. [Epub ahead of print]. [Crossref] [PubMed]
- Tang D, Yang C, Zheng J, et al. Image-based modeling and precision medicine: patient-specific carotid and coronary plaque assessment and predictions. IEEE Trans Biomed Eng 2013;60:643-51. [Crossref] [PubMed]
- Bede P, Hardiman O. Lessons of ALS imaging: Pitfalls and future directions - A critical review. Neuroimage Clin 2014;4:436-43. [Crossref] [PubMed]
- Tan CC, Yu JT, Tan L. Biomarkers for preclinical Alzheimer's disease. J Alzheimers Dis 2014;42:1051-69. [PubMed]
- Jiang T, Yu JT, Tan L. Novel disease-modifying therapies for Alzheimer's disease. J Alzheimers Dis 2012;31:475-92. [PubMed]
- Mills PB, Struys E, Jakobs C, et al. Mutations in antiquitin in individuals with pyridoxine-dependent seizures. Nat Med 2006;12:307-9. [Crossref] [PubMed]


