
A deep learning-based hybrid artificial intelligence model for the detection and severity assessment of vitiligo lesions
Introduction
Vitiligo is a common, acquired, patchy, depigmented skin disease that can be localized or widespread. The prevalence of vitiligo varies worldwide, with previous studies reporting a rate of 0.2% to 1.8% (1,2). Despite its benign nature, the burden of vitiligo is comparable to other skin diseases such as psoriasis and eczema (3). Observable lesions, especially those on readily visible sites, such as the face, neck, and hands, can lead to shame, anxiety, and depression (4). The chronicity of vitiligo and the lack of satisfactory treatments may significantly reduce patients’ quality of life and cause substantial psychosocial stress (3-7).
Diagnosis of vitiligo is often straightforward, but assessment of the disease is challenging, as lesions vary greatly in number, size, location, morphology, and extent of depigmentation. Repigmentation, either spontaneous or treatment-induced, may also take different forms and is often unevenly distributed (8,9). Assessment of changes in the size and pigmentation of lesions are indispensable for tracking the course of vitiligo, determining the disease progression, evaluating treatment outcomes as well as comparing different treatment approaches in observational studies or prospective clinical trials. Nevertheless, valid, accessible, and user-friendly assessment tools are lacking. To date, several methods for vitiligo assessment have been proposed. Subjective and semi-objective methods are either empirical or based on clinical scales such as the Vitiligo European Task Force assessment (VETFa) and the Vitiligo Area Scoring Index (VASI) (10-14). These methods are limited by their lack of objectivity and consistency and the complexity of their application in routine clinical settings (15-17). Objective methods, such as colorimetry, reflectance confocal microscopy (RCM), and image analysis, represent a growing trend of digital evaluation (18-24). These procedures focus on size measurement or color changes of vitiligo lesions and often require special equipment, expertise, or laborious input, which affects their availability. A comprehensive assessment tool covering both morphometry and colorimetry has yet to be developed.
In recent years, the deep learning method of artificial intelligence (AI) has been used extensively in medical image analysis and other diverse clinical scenarios. Unlike previous machine learning methods, deep learning uses complex, multilayer, artificial neural networks to achieve pattern recognition, data classification, prediction, and decision-making support on a massive scale (25). In dermatology, Esteva et al. first reported a deep convolutional neural network (DCNN) that successfully classified melanoma and keratinocyte cancers at the same level of competency as a board-certified dermatologist (26). Recent DCNN architectures have further demonstrated a comparable or even superior competence to dermatologists in disease classification, differentiation, and dermatopathological analysis (27-30). For vitiligo assessment, DCNN may potentially extract both morphometric and colorimetric features at a pixel level, which would enable it to detect and outline vitiligo lesions with similar efficiency but exceeding reproductivity and consistency, overcoming the limitations of existing methods. Several previous studies reported the use in vitiligo assessment of traditional classifier models such as support vector machines (SVMs) and k-nearest neighbor (KNN) algorithms (31-33). However, the application of DCNN is limited, and DCNN-based assessment tools are lacking. Therefore, the present study aimed to construct and validate a DCNN-based hybrid AI model for the morphometric and colorimetric evaluation of vitiligo lesions which might contribute considerably to a speedy and reliable disease assessment in diverse clinical, research and development scenarios. We present the following article in accordance with the TRIPOD reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1738/rc).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the medical ethics committee of Hospital for Skin Disease and Institute of Dermatology, Chinese Academy of Medical Sciences (CAMS) & Peking Union Medical College [Approval No. 2017 (022)]. Informed consent was taken from all the patients. We adopted a hybrid AI design, using 2 DCNNs sequentially to achieve classification/localization and segmentation of vitiligo lesions, respectively. This AI model was trained, fine-tuned, and tested followed by 3 add-on metrics for automated size measurement and pigmentation assessments in comparison with dermatologist evaluators (Figure 1).
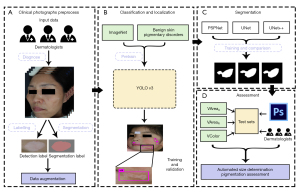
Datasets
The dataset used in this study was mainly retrieved from the image database of the Department of Cosmetic Laser Surgery in the Hospital for Skin Disease and Institute of Dermatology, Chinese Academy of Medical Sciences (CAMS) & Peking Union Medical College. Clinical photographs of vitiligo lesions were taken from patients with Fitzpatrick skin types III or IV who visited the department from 2003 to 2019 using digital single-lens reflex (DSLR) cameras (EOS 550D/800D, Canon, Tokyo, Japan; FinePix S9500, Fujifilm, Tokyo, Japan). These photographs were numbered based on the patient’s unique identification number assigned during his or her first visit. Images of insufficient quality (e.g., blurry, underexposed, or overexposed images) were removed from the dataset.
The classification/localization study included a total of 2,720 images, which were divided into a training set (80%) and a validation set (16%). These images were diagnosed as vitiligo by 3 dermatologists in consensus, and the lesions were further labeled with ground truth values using LabelImg (https://github.com/tzutalin/labelImg). Images underwent flipping and random rotation augmentation prior to training. A test set was set up that consisted of 100 (4%) images containing 167 vitiligo lesions and 100 control images of normal skin. The images in the 3 different sets were nonoverlapping and completely independent from one another (Figure 2).
The segmentation study included a total of 1,262 images of vitiligo lesions, which were divided into a training set (80%), a validation set (10%), and a test set (10%). These images were initially segmented using nEO iMAGING version 4.4.1 (NEO iMAGING Lab, Shenzhen, China) by 1 dermatologist and further adjusted by 2 other dermatologists to reach a consensus. Images were preprocessed before training as mentioned above. The images in the 3 different sets were nonoverlapping and independent from one another (Figure 2).
Additionally, to test the model’s performance on lesions of Fitzpatrick skin types I, II, and V, we included another test set of 145 vitiligo lesions accessed from online public databases.
The test set consisted of 20 DSLR-acquired images containing up to 27 lesions from 20 patients, each with a 1 cm by 2 cm scale tag, and 20 pairs of VISIA®-CR (Canfield Imaging Systems, Fairfield, NJ, USA)-acquired images taken before and after 308 nm XeCl excimer laser (XTRAC, PhotoMedex, Montgomeryville, PA, USA) treatment, which were used to test the AI model’s measurement of the area and pigmentation of vitiligo lesions, respectively (Figure 2).
DCNN training and add-on metrics
A 3-stage hybrid AI model was constructed (Figure 1). First, to classify and locate vitiligo lesions, we used the YOLO v3 architecture pretrained on ImageNet and benign skin pigmentary disorders (unpublished data). Next, 3 DCNNs, namely Pyramid Scene Parsing Network (PSPNet), UNet, and UNet++, were trained and compared for their ability to segment vitiligo lesions. The architecture with the best performance was integrated into the AI model. The model was trained and tested on a server (Intel Xeon CPU E5 @ 2.10 GHz, 32 GB RAM, 1080 GTX GPU). The hyperparameters of the YOLO v3, PSPNet, UNet, and UNet++ architectures trained in this study are shown in Tables S1,S2.
Finally, 3 add-on metrics, VAreaA, VAreaR, and VColor were attached to the trained DCNNs to perform morphometric and colorimetric assessments.
VAreaA represented the absolute size of a lesion. With reference to the scale tag, area (cm2) per pixel was calculated, and the area of a lesion was determined based on the pixel to area transformation.
VAreaR represented the relative size change of a lesion by comparing the total pixels of matched pre- and post-treatment lesions. VAreaR was defined as follows: . VAreaR ranged from 0 to 1 (0–100%), with higher values signifying a greater reduction in lesion area.
VColor measured the relative pigmentation changes of a given lesion based on IWANewtone. IWANewtone is an index proposed by Newtone Technologies to assess skin whiteness that takes into account both melanin and hemoglobin (34). IWANewtone was defined as follows: , where L*, a*, and b* were the 3 color components of the system proposed by the International Commission of Lighting (CIE) (35). A higher IWANewtone indicated a whiter skin color. The RGB color space of matched pre- and post-treatment lesions, as well as normal skin, was converted into the CIE L*a*b* color space, and IWANewtone was thus calculated. VColor was defined as follows:. .VColor ranged from 0 to 1 (0–100%), with higher values signifying a greater level of repigmentation.
Evaluation of the AI model and statistical analysis
The performance of the YOLO v3 architecture was evaluated on the test set in reference to the dermatologists’ consensus annotation, with sensitivity and error rate as primary outcomes.
The Jaccard index (JI), also known as the intersection over union metric (IoU), was used to assess the segmentation performance of the PSPNet, UNet, and UNet++ architectures. In the context of the present study, JI represented the area of overlap between the DCNN’s and the dermatologists’ segmentation divided by the area of their union. This outcome ranged from 0–1 (0–100%), with higher values signifying a better segmentation.
To evaluate the model’s ability to measure the area of vitiligo lesions, the VAreaA data of a test set consisting of 20 DSLR-acquired, scaled images were calculated in comparison with Adobe Photoshop 7 (Adobe, San Jose, CA, USA)-derived area data obtained by a trained dermatologist as previously described (24). A Wilcoxon rank sum test was performed to assess the difference between the AI model and the Photoshop results.
VAreaR data were obtained on a test set of 20 pairs of VISIA®-CR-acquired images in comparison with Photoshop-derived data and an independent assessment by 3 trained dermatologists using a 5-grade Physician’s Global Assessment (PGA) for visual inspection. A paired t-test was conducted to assess the difference between the AI model and the Photoshop results. The VAreaR data were further categorized into the 5 PGA grades (0, Grade 0; 1–5%, Grade 1; 26–50%, Grade 2; 51–75%, Grade 3; 76–100%, Grade 4). Kendall’s W test was used to assess the degree of agreement among the AI model, Photoshop, and the dermatologists and the concordance between the dermatologists.
To evaluate the model’s ability to detect the colorimetric changes of vitiligo lesions, VColor data were obtained from the same test set as VAreaR and compared with the consensus results of 3 trained dermatologists who graded the residual depigmentation of lesions before and after treatment according to VASI (10). Both the VColor and the VASI data were converted into a 5-grade scale following PGA and analyzed using the weighted kappa coefficient test.
Descriptive statistical measures, such as mean, standard deviation (SD), frequency, and range were used. All statistical analyses were conducted using SPSS 24 (IBM Corp., Armonk, NY, USA). Results were regarded as statistically significant when P<0.05.
Results
In a test set of 167 lesions and 100 controls, the YOLO v3 architecture correctly identified 155 lesions (sensitivity 92.91%), with an overall error rate of 14.98% (40/267). Twenty-eight cases (10.41%) were wrongly labeled. Among these, 16 cases of light reflections and 6 cases of light-pigmented facial parts were misclassified as vitiligo lesions. In addition, the architecture failed to detect 12 cases (7.19%) of vitiligo lesions, most of which were either small in size (6/12, 50%) or located on the scalp or near the mouth (3/12, 25%). For the segmentation of vitiligo lesions, the UNet++ architecture showed the best performance, with a JI of 0.79 in comparison with the PSPNet (JI: 0.746) and UNet (JI: 0.706) architectures (P<0.05; Figure 3). Consequently, UNet++ was integrated into the model. On the additional test set, however, the model achieved a lower detection sensitivity of 72.41% (105/145), as well as a lower segmentation JI of 0.69.
The results of the absolute size measurements are shown in Table 1. On a test of 20 images with 27 lesions, the median VAreaA was 1.2437 cm2, while the median area obtained by the Photoshop analysis was 1.3114 cm2. No statistical difference was observed between the AI model and the Photoshop analysis (P=0.075).
Table 1
| Test No. | VAreaA (cm2) | Photoshop (cm2) |
|---|---|---|
| 1 | 7.1147 | 7.5472 |
| 2 | 1.7613 | 1.5030 |
| 3 | 1.6227 | 1.4489 |
| 4 | 4.4137 | 4.9541 |
| 5 | 6.5187 | 7.2142 |
| 6 | 1.3207 | 1.4562 |
| 7 | 0.9003 | 0.9587 |
| 8 | 1.8766 | 2.1007 |
| 9 | 18.6896 | 19.6916 |
| 10 | 0.2173 | 0.1977 |
| 11 | 0.1084 | 0.1255 |
| 12 | 0.8499 | 0.9528 |
| 13 | 0.8649 | 0.9433 |
| 14 | 1.7278 | 1.6236 |
| 15 | 0.7549 | 0.8153 |
| 16 | 0.1311 | 0.1316 |
| 17 | 1.6279 | 1.7883 |
| 18 | 0.5641 | 0.46228 |
| 19 | 0.464 | 0.3752 |
| 20 | 1.3851 | 1.3114 |
| 21 | 1.4994 | 1.6747 |
| 22 | 1.2437 | 1.4337 |
| 23 | 0.1515 | 0.1654 |
| 24 | 1.1069 | 0.8556 |
| 25 | 0.1164 | 0.1239 |
| 26 | 1.6629 | 1.8054 |
| 27 | 0.1245 | 0.1292 |
VAreaA, a metric to evaluate absolute lesion size.
The results of the relative size measurements are shown in Table 2. The mean VAreaR of the test set samples was 83.15% (SD: 22.74%), while the mean relative area obtained by the Photoshop analysis was 80.91% (SD: 23.18%). No significant difference was found between the AI model and the Photoshop analysis (P=0.212). Kendall’s W test showed a high level of concordance between the AI model and the dermatologists (W=0.812, P<0.05). Also, a high degree of agreement was found between the Photoshop analysis and the dermatologists (W=0.857, P<0.05) as well as between the 3 dermatologists themselves (W=0.858, P<0.05).
Table 2
| Test No. | (%/Grade†) | ||||
|---|---|---|---|---|---|
| VAreaR | PS | D1 | D2 | D3 | |
| 1 | 51.30/3 | 40.75/2 | 50/2 | 35/2 | 50/2 |
| 2 | 100.00/4 | 100/4 | 100/4 | 90/4 | 95/4 |
| 3 | 73.24/3 | 84.27/4 | 80/4 | 90/4 | 80/4 |
| 4 | 42.08/2 | 33.6/2 | 30/2 | 30/2 | 15/1 |
| 5 | 19.01/1 | 29.84/2 | 20/1 | 30/2 | 10/1 |
| 6 | 95.00/4 | 85.15/4 | 90/4 | 70/3 | 90/4 |
| 7 | 93.21/4 | 81.62/4 | 90/4 | 90/4 | 80/4 |
| 8 | 62.72/3 | 61.04/3 | 50/2 | 50/2 | 60/3 |
| 9 | 96.66/4 | 95.82/4 | 90/4 | 90/4 | 95/4 |
| 10 | 100.00/4 | 100/4 | 100/4 | 90/4 | 95/4 |
| 11 | 98.61/4 | 100/4 | 100/4 | 80/4 | 95/4 |
| 12 | 86.90/4 | 80.18/4 | 80/4 | 60/3 | 70/3 |
| 13 | 90.51/4 | 100/4 | 90/4 | 100/4 | 100/4 |
| 14 | 99.79/4 | 100/4 | 100/4 | 80/4 | 98/4 |
| 15 | 89.45/4 | 80.08/4 | 80/4 | 75/3 | 60/3 |
| 16 | 100.00/4 | 100/4 | 100/4 | 100/4 | 100/4 |
| 17 | 100.00/4 | 91.76/4 | 90/4 | 85/4 | 70/3 |
| 18 | 78.97/4 | 62.85/3 | 80/4 | 60/3 | 70/3 |
| 19 | 85.51/4 | 91.21/4 | 80/4 | 90/4 | 80/4 |
| 20 | 100.00/4 | 100/4 | 100/4 | 95/4 | 85/4 |
†, 5-grade PGA. VAreaA, a metric to evaluate absolute lesion size; PS, Photoshop; D1, Dermatologist 1; D2, Dermatologist 2; D3, Dermatologist 3; PGA, Physician’s Global Assessment.
The results of the colorimetric assessment are shown in Table 3. A good level of agreement between the VColor and the dermatologists’ VASI data was seen (κ=0.869, P<0.05).
Table 3
| Test No. | (%/Grade†) | |
|---|---|---|
| VColor | VASI‡ | |
| 1 | 76.92/4 | 50/1 |
| 2 | 44.44/1 | 50/1 |
| 3 | 0/0 | 0/0 |
| 4 | 42.86/1 | 65/2 |
| 5 | 100/4 | 80/4 |
| 6 | 81.82/4 | 75/3 |
| 7 | 96.15/4 | 90/4 |
| 8 | 81.25/4 | 80/4 |
| 9 | 25/0 | 40/1 |
| 10 | 40.91/1 | 65/2 |
| 11 | 50/1 | 65/2 |
| 12 | 66.67/2 | 65/2 |
| 13 | 85.71/4 | 90/4 |
| 14 | 7.69/0 | 15/0 |
| 15 | 100/4 | 90/4 |
| 16 | 73.91/3 | 90/4 |
| 17 | 36.67/2 | 50/2 |
| 18 | 43.48/2 | 50/2 |
| 19 | 85.71/4 | 90/4 |
| 20 | 31.82/2 | 50/2 |
†, 5-grade PGA; ‡, consensus results of 3 trained dermatologists. VColor, a metric to evaluate colorimetric (pigmentation) changes; VASI, Vitiligo Area Scoring Index; PGA, Physician’s Global Assessment.
Discussion
Although the use of deep learning in dermatology has surged during the past few years, most of this work has focused on pigmented skin neoplasms. For depigmented skin conditions, such as vitiligo, limited data are available. Two earlier studies reported deep learning models that achieved a sensitivity of 92% and 86%, respectively, for the classification of vitiligo (36,37). However, both studies were trained on small sample sets and are yet unvalidated. In 2019, Liu et al. investigated 4 common DCNNs (Resnet50, Vgg16, Xception, and Iception v3), which were trained on the same vitiligo dataset with 3 different color space images (RGB, HSV, and YCrCb) (38). In this study, the “double” combination (combined DCNNs and combined color space image processing methods) showed the best performance, with an accuracy of 87.8%, a precision of 91.9%, a sensitivity of 90.9%, and a specificity of 80.2% (38). Li et al. proposed an image synthesis algorithm. A convolutional neural network (CNN) modified by a fully convolutional network (FCN) was trained using synthetic and internet images to segment vitiligo lesions, and the proposed algorithm had an error of 1.06% for facial vitiligo area estimation (39). More recently, Liu et al. presented an Inception v4-based deep learning system for the differential diagnosis of 26 common skin diseases (30). Upon validation with clinicians to classify vitiligo lesions, this system yielded a sensitivity comparable to dermatologists and superior to primary care physicians and nurse practitioners. In the present study, we used the YOLO v3 architecture for classification. The sensitivity of our model was 92.91%, indicating a good capacity to classify vitiligo lesions. Moreover, this DCNN simultaneously classified and located vitiligo lesions in 1 step and showed advantages in detecting small objects, which is valuable in the context of the varying numbers and sizes of vitiligo lesions and may significantly facilitate subsequent segmentation tasks. However, it should be noted that this model mislabeled several reflections as vitiligo lesions, warranting further image processing and model tuning to address this limitation.
Lesion segmentation, namely separating lesions from surrounding normal skin, is a critical step in the evaluation of skin diseases. Although several segmentation methods for vitiligo have been reported (32,40-42), the DCNN-based approach is scarce in the literature. In the present study, we trained and validated the UNet++ architecture, which is a powerful DCNN for diverse medical image segmentation tasks. It achieved a JI of 0.79, which was better than that of UNet and PSPNet when compared with dermatologists’ manual segmentation. Recently, Low et al. reported a modified UNet architecture that used a ResNet50 contracting path for the segmentation of vitiligo lesions (43). Although trained on a small sample set, it scored 0.815, a higher JI than that of our model. It also showed a superior segmentation to the original UNet, especially on lesions of complex morphology. Our future work may thus explore and fuse together modalities with the best performance from different DCNNs to enhance the segmentation capacity of the AI model.
In recent years, morphometry-based assessments, such as Photoshop, Image Pro Plus, and AutoCAD2000 have been shown to provide precise and accessible evaluations of vitiligo lesions (20-22,24). However, the use of these analysis tools tends to be time-consuming and skill dependent. AI-based evaluation may overcome these disadvantages. For the assessment of absolute (VAreaA) and relative (VAreaR) size changes, our AI model yielded results consistent with those of Photoshop analysis and trained dermatologists. Compared with other image analysis methods, this AI model may facilitate the measurement of vitiligo lesions due to instant localization and segmentation. Despite these promising results, the size assessment of this model was performed in a limited way, and more validation work is needed.
Pigmentation evaluation is challenging, as visual perception of different shades of skin color is subjective and can differ widely from one individual to another. So far there has been no universally accepted metric to assess skin pigmentation in vitiligo research. Colorimetry and RCM measure skin pigmentation quantitatively and provide precise data (18-20).However, these methods are expertise-demanding, not readily available, and may function poorly on lesions with uneven pigmentation. VETF is a staging system based on cutaneous and hair pigmentation that divides vitiligo lesions into 3 less well-defined stages (11). VASI is another validated score system offering more accurate measures of pigmentation by grading the lesions according to the percentage of pigment loss (12). VASI generates 7 gradient scores, 0, 10%, 25%, 50%, 75%, 90%, or 100%, with higher percentages signifying a higher degree of depigmentation. In our study, we designed a percentage metric, VColor, to represent the change of pigmentation. VColor was based on IWANewtone, an index of skin tone that is measured continuously, rendering VColor more sensitive to pigmentation changes than VETFa or VASI. To allow for direct comparison, we converted both VColor and VASI data into a 5-grade PGA scale, and the results showed that VColor agreed well with the VASI assessment performed by the dermatologists. This metric was more objective than VASI assessment, and it was calculated directly on clinical images, thus making it easier to conduct than other equipment-based colorimetric analysis. However, it is important to note that light conditions may significantly affect the IWANewtone. Therefore, VColor measurement is best performed on images acquired with VISIA or similar imaging systems with standardized illuminations and fixed camera systems.
A major limitation of this study was the lack of ethnic skin types other than Fitzpatrick skin types III and IV in the training data. Unsurprisingly, the model showed a lower detection sensitivity and segmentation accuracy when assessing vitiligo lesions of Fitzpatrick skin types I, II, or V. Further research using data from a wider variety of sources would improve the performance of the model on various ethnic skin groups and extend its usability.
Conclusions
We established and validated a novel hybrid AI model for the objective evaluation of vitiligo lesions from Asian patients with Fitzpatrick skin types III or IV. Using YOLO v3 and UNet++ architectures sequentially, this model achieved lesion classification, localization, and segmentation. Three add-on metrics, VAreaA, VAreaR, and VColor, demonstrated comparable measurements with image-analysis software and trained dermatologists in terms of both morphometric and colorimetric assessment of vitiligo lesions. Using clinical images only, this model is accessible and suitable for various clinical settings.
Acknowledgments
We are grateful to Nanjing Suoyousuoyi Information Technology Co., Ltd. and Nanjing Hongtu Institute of Artificial Intelligence Technology for assisting in building the models.
Funding: This work was supported by the Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (No. CIFMS-2021-12M-001).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1738/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1738/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1738/coif). All authors report technical assistance from Nanjing Suoyousuoyi Information Technology Co., Ltd. and Nanjing Hongtu Institute of Artificial Intelligence Technology. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the medical ethics committee of Hospital for Skin Disease and Institute of Dermatology, Chinese Academy of Medical Sciences (CAMS) & Peking Union Medical College [Approval No. 2017 (022)]. Informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhang Y, Cai Y, Shi M, et al. The prevalence of vitiligo: A meta-analysis. PLoS One 2016;11:e0163806. [Crossref] [PubMed]
- Zhang JZ, Luo D, An CX, et al. Clinical and epidemiological characteristics of vitiligo at different ages: An analysis of 571 patients in Northwest China. International Journal of Dermatology and Venereology 2019;2:165-8. [Crossref]
- Linthorst Homan MW, Spuls PI, de Korte J, et al. The burden of vitiligo: patient characteristics associated with quality of life. J Am Acad Dermatol 2009;61:411-20. [Crossref] [PubMed]
- Salzes C, Abadie S, Seneschal J, et al. The Vitiligo Impact Patient Scale (VIPs): Development and validation of a vitiligo burden assessment tool. J Invest Dermatol 2016;136:52-8. [Crossref] [PubMed]
- Elbuluk N, Ezzedine K. Quality of life, burden of disease, co-morbidities, and systemic effects in vitiligo patients. Dermatol Clin 2017;35:117-28. [Crossref] [PubMed]
- Ezzedine K, Eleftheriadou V, Whitton M, et al. Vitiligo. Lancet 2015;386:74-84. [Crossref] [PubMed]
- Chen CY, Wang WM, Chung CH, et al. Increased risk of psychiatric disorders in adult patients with vitiligo: A nationwide, population-based cohort study in Taiwan. J Dermatol 2020;47:470-5. [Crossref] [PubMed]
- Gan EY, Eleftheriadou V, Esmat S, et al. Repigmentation in vitiligo: position paper of the Vitiligo Global Issues Consensus Conference. Pigment Cell Melanoma Res 2017;30:28-40. [Crossref] [PubMed]
- Bergqvist C, Ezzedine K. Vitiligo: A focus on pathogenesis and its therapeutic implications. J Dermatol 2021;48:252-70. [Crossref] [PubMed]
- Hamzavi I, Jain H, McLean D, et al. Parametric modeling of narrowband UV-B phototherapy for vitiligo using a novel quantitative tool: the Vitiligo Area Scoring Index. Arch Dermatol 2004;140:677-83. [Crossref] [PubMed]
- Taïeb A, Picardo M. VETF Members. The definition and assessment of vitiligo: a consensus report of the Vitiligo European Task Force. Pigment Cell Res 2007;20:27-35. [Crossref] [PubMed]
- Komen L, da Graça V, Wolkerstorfer A, et al. Vitiligo Area Scoring Index and Vitiligo European Task Force assessment: reliable and responsive instruments to measure the degree of depigmentation in vitiligo. Br J Dermatol 2015;172:437-43. [Crossref] [PubMed]
- van Geel N, Bekkenk M, Lommerts JE, et al. The Vitiligo Extent Score (VES) and the VESplus are responsive instruments to assess global and regional treatment response in patients with vitiligo. J Am Acad Dermatol 2018;79:369-71. [Crossref] [PubMed]
- Mogawer RM, Mostafa WZ, Elmasry MF. Comparative analysis of the body surface area calculation method used in vitiligo extent score vs the hand unit method used in vitiligo area severity index. J Cosmet Dermatol 2020;19:2679-83. [Crossref] [PubMed]
- Alghamdi KM, Kumar A, Taïeb A, et al. Assessment methods for the evaluation of vitiligo. J Eur Acad Dermatol Venereol 2012;26:1463-71. [Crossref] [PubMed]
- Eleftheriadou V, Thomas KS, Whitton ME, et al. Which outcomes should we measure in vitiligo? Results of a systematic review and a survey among patients and clinicians on outcomes in vitiligo trials. Br J Dermatol 2012;167:804-14. [Crossref] [PubMed]
- Mikhael NW, Sabry HH, El-Refaey AM, et al. Clinimetric analysis of recently applied quantitative tools in evaluation of vitiligo treatment. Indian J Dermatol Venereol Leprol 2019;85:466-74. [Crossref] [PubMed]
- Park SB, Suh DH, Youn JI. A long-term time course of colorimetric evaluation of ultraviolet light-induced skin reactions. Clin Exp Dermatol 1999;24:315-20. [Crossref] [PubMed]
- Van Geel N, Vander Haeghen Y, Ongenae K, et al. A new digital image analysis system useful for surface assessment of vitiligo lesions in transplantation studies. Eur J Dermatol 2004;14:150-5. [PubMed]
- Longo C, Bassoli S, Farnetani F, et al. Reflectance confocal microscopy for melanoma and melanocytic lesion assessment. Expert Rev Dermatol 2008;3:735-45. [Crossref]
- Marrakchi S, Bouassida S, Meziou TJ, et al. An objective method for the assessment of vitiligo treatment. Pigment Cell Melanoma Res 2008;21:106-7. [Crossref] [PubMed]
- Welsh O, Herz-Ruelas ME, Gómez M, et al. Therapeutic evaluation of UVB-targeted phototherapy in vitiligo that affects less than 10% of the body surface area. Int J Dermatol 2009;48:529-34. [Crossref] [PubMed]
- Kang HY, Bahadoran P, Ortonne JP. Reflectance confocal microscopy for pigmentary disorders. Exp Dermatol 2010;19:233-9. [Crossref] [PubMed]
- El-Zawahry MB, Zaki NS, Wissa MY, et al. Effect of combination of fractional CO2 laser and narrow-band ultraviolet B versus narrow-band ultraviolet B in the treatment of non-segmental vitiligo. Lasers Med Sci 2017;32:1953-8. [Crossref] [PubMed]
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436-44. [Crossref] [PubMed]
- Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115-8. [Crossref] [PubMed]
- Haenssle HA, Fink C, Schneiderbauer R, et al. Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann Oncol 2018;29:1836-42. [Crossref] [PubMed]
- Andres C, Andres-Belloni B, Hein R, et al. iDermatoPath - a novel software tool for mitosis detection in H&E-stained tissue sections of malignant melanoma. J Eur Acad Dermatol Venereol 2017;31:1137-47. [Crossref] [PubMed]
- Olsen TG, Jackson BH, Feeser TA, et al. Diagnostic performance of deep learning algorithms applied to three common diagnoses in dermatopathology. J Pathol Inform 2018;9:32. [Crossref] [PubMed]
- Liu Y, Jain A, Eng C, et al. A deep learning system for differential diagnosis of skin diseases. Nat Med 2020;26:900-8. [Crossref] [PubMed]
- Arifin MS, Kibria MG, Firoze A, et al. editors. Dermatological disease diagnosis using color-skin images. 2012 international conference on machine learning and cybernetics; 2012; IEEE.
- Shamsudin N, Hussein SH, Nugroho H, et al. Objective assessment of vitiligo with a computerised digital imaging analysis system. Australas J Dermatol 2015;56:285-9. [Crossref] [PubMed]
- Nosseir A, Shawky MA. editors. Automatic classifier for skin disease using k-NN and SVM. Proceedings of the 2019 8th international conference on software and information engineering; 2019.
- Martin‐Langrand G, Beaufrère‐Seron B, Korichi R, et al. Facial and Skin Radiance: New Indexes. New York, NY: IFSCC; 2014.
- Weatherall IL, Coombs BD. Skin color measurements in terms of CIELAB color space values. J Invest Dermatol 1992;99:468-73. [Crossref] [PubMed]
- Yasir R, Rahman MA, Ahmed N. editors. Dermatological disease detection using image processing and artificial neural network. 8th International Conference on Electrical and Computer Engineering; 2014; IEEE.
- Anthal J, Upadhyay A, Gupta A. editors. Detection of vitiligo skin disease using LVQ neural network. 2017 International Conference on Current Trends in Computer, Electrical, Electronics and Communication (CTCEEC); 2017; IEEE.
- Liu J, Yan J, Chen J, et al. editors. Classification of vitiligo based on convolutional neural network. International Conference on Artificial Intelligence and Security; 2019; Springer.
- Li Y, Kong AW, Thng S. Segmenting vitiligo on clinical face images using CNN Trained on Synthetic and Internet Images. IEEE J Biomed Health Inform 2021;25:3082-93. [Crossref] [PubMed]
- Nugroho H, Fadzil MH, Yap VV, et al. Determination of skin repigmentation progression. Annu Int Conf IEEE Eng Med Biol Soc 2007;2007:3442-5. [PubMed]
- Sheth VM, Rithe R, Pandya AG, et al. A pilot study to determine vitiligo target size using a computer-based image analysis program. J Am Acad Dermatol 2015;73:342-5. [Crossref] [PubMed]
- Chica JF, Zaputt S, Encalada J, et al. Objective assessment of skin repigmentation using a multilayer perceptron. J Med Signals Sens 2019;9:88-99. [Crossref] [PubMed]
- Low M, Huang V, Raina P. editors. Automating vitiligo skin lesion segmentation using convolutional neural networks. 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI); 2020; IEEE.
(English Language Editor: C. Gourlay)




