Mining of single nucleotide polymorphisms in the 3' untranslated region of liver cancer-implicated miR-122 target genes
Currently there has been a lot of focus on the microRNA (miR)-122 to understand the genetics of hepatocellular carcinoma (HCC), pathology of hepatitis B virus (HBV) and hepatitis C virus (HCV) infections, hepatic insulin resistance, and lipid metabolism (1-4). Interestingly, miR-122 expression levels were significantly reduced than the normal levels in HBV-associated liver cancer, but not in HCV related liver cancer (5). Also, there is an overwhelming amount of data suggesting the role of single nucleotide polymorphisms (SNPs) in hepatic genes and its association with the altered risk/development of hepatic cancer and its progression. Polymorphisms in the miRNA-binding sites of the target genes are more frequent than SNPs in miRNA genes and therefore, it is considered that polymorphisms in the cytokines and other genes have correlations with chronic HBV or HCV infections. These SNPs in the miRNA binding sites of the target genes can potentially enhance or weaken the interaction between the miRNAs and the target transcripts. Therefore, it is important to study the SNPs in miR-122 binding sites of the target genes to understand the genetic basis of HCC.
Tumor suppressor miR-122 levels are relatively different in both HBV and HCV infections although both types of infection ultimately can lead to HCC. This is an interesting evidence to look for alternate mechanisms involved in hepatic carcinogenesis. The use of in silico prediction and experimental validation will only be the beginning steps in a large scale effort to analyze a variety of clinical liver cancer samples associated with altered miR-122 levels. Therefore, in this report by using bioinformatics tool we catalogued SNPs in the 3' untranslated region (UTR) of hepatic cancer implicated genes that can affect miR-122 regulation (1, 6). The list of genes in the current study was chosen based on the previous study (1). These genes have miR-122 binding sites and also participate in pathogenesis of HCC as reported by Tsai et al., 2009 (1). Analyses of SNPs and INDELs in miR-122 target sites were performed by using PolymiRTS Database 3.0 that can be accessed at http://compbio.uthsc.edu/miRSNP/ (6). The Percentage of SNPs in 3' UTR of all the studied genes were obtained by using dbSNP database that can be accessed at https://www.ncbi.nlm.nih.gov/snp. The results are shown in Table 1. The table provides information about SNPs/INDELs in miR-122 target sites of hepatic cancer implicated genes that are predicted by the PolymiRTS database.
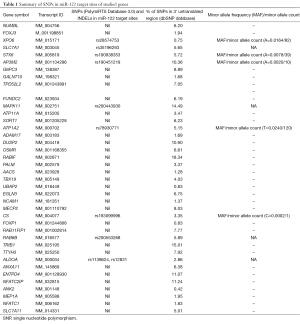
Full table
We believe that our report provides the first step towards integrating SNP analysis with studies on miRNA-122 and global de-repression of host miR-122 targets in hepatic cancer cells. This will provide a suitable base for future research that can improve our understanding of 3' UTR polymorphisms and the failure of miR122 regulation in varied clinical samples. Therefore, integrating SNP analysis with studies on miR-122 regulation in liver cancer cells by framing suitable hypotheses and experimental designs can result in the development of novel cancer-targeting therapeutics.
Acknowledgements
PS Suresh would like to thank Faculty Recharge Programme of UGC for the support.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Tsai WC, Hsu PW, Lai TC, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology 2009;49:1571-82. [Crossref] [PubMed]
- Yang YM, Seo SY, Kim TH, et al. Decrease of microRNA-122 causes hepatic insulin resistance by inducing protein tyrosine phosphatase 1B, which is reversed by licorice flavonoid. Hepatology 2012;56:2209-20. [Crossref] [PubMed]
- Li C, Wang Y, Wang S, et al. Hepatitis B virus mRNA-mediated miR-122 inhibition upregulates PTTG1-binding protein, which promotes hepatocellular carcinoma tumor growth and cell invasion. J Virol 2013;87:2193-205. [Crossref] [PubMed]
- Luna JM, Scheel TK, Danino T, et al. Hepatitis C virus RNA functionally sequesters miR-122. Cell 2015;160:1099-110. [Crossref] [PubMed]
- Spaniel C, Honda M, Selitsky SR, et al. microRNA-122 abundance in hepatocellular carcinoma and non-tumor liver tissue from Japanese patients with persistent HCV versus HBV infection. PLoS One 2013;8:e76867. [Crossref] [PubMed]
- Bhattacharya A, Ziebarth JD, Cui Y. PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res 2014;42:D86-91. [Crossref] [PubMed]


