
A comprehensive characterization of alternative splicing events related to prognosis and the tumor microenvironment in lung adenocarcinoma
Introduction
Currently, lung cancer is the leading cause of cancer incidence and cancer-related mortality in China (1) and in the world (2). Non-small cell lung cancer (NSCLC) accounts for approximately 85% of lung cancer diagnoses, and can be further subdivided into lung adenocarcinoma (LUAD), squamous cell carcinoma, and large cell carcinoma (3). LUAD is the most prevalent subtype and accounts for 50% of NSCLC (4). Although many opportunities exist for improving the treatment of LUAD, including novel immunotherapy, molecular targets, and antiangiogenic therapies, the prognosis of patients with LUAD is often poor due to late diagnosis, tumor heterogeneity, and a lack of understanding of the disease mechanisms. Indeed, the 5-year survival rate is approximately 15% (1,2). The lack of accurate clinical classification and prognostic assessment system is currently a major clinical problem in LUAD. Therefore, there is an urgent need to develop sensitive and effective biomarkers for diagnosis and prognosis prediction. Notably, the role of alternative splicing (AS) in LUAD prognosis and therapy prediction has been increasingly recognized (5).
AS is an essential mechanism in regulating biological systems, generating multiple messenger-RNA (mRNA) and protein isoforms, and is regulated by alternative splicing factors (SFs) (6). A previous study has indicated that more than 95% of human genes experience AS in physiological processes (7). Dysregulation of SFs can promote the emergence of numerous aberrant AS events in cancer (8,9), which may play an important role in driving the tumorigenesis and progression of malignancies by facilitating cell proliferation, apoptosis, invasion/metastasis, angiogenesis, and immune escape (10,11). Aberrant AS events are also believed to affect the effectiveness of cancer chemotherapy, targeted therapy, and/or immunotherapy (12,13). Exploration of aberrant AS events will help us better understand the underlying mechanisms of LUAD initiation and development, so as to further guide clinical practice. Furthermore, AS has emerged as a key event in the formation of the tumor microenvironment (TME) (14,15).
The TME is a complex system containing endothelial cells, fibroblasts, pericytes, structural components, and infiltrating immune cells (16,17), all of which impact tumor development, invasion, and metastasis (18). Emerging evidence has demonstrated that the TME components can modify the immune phenotypes of tumors and therefore affect the patient’s prognosis (19-21). For instance, cancer associated fibroblasts play a direct immunosuppressive mechanism (21), whereas cytotoxic immune cells promote antitumor immune responses (19). Due to the important role of the TME in malignancies, assessment of the heterogeneity of the TME may effectively predict therapeutic benefits and prognosis for patients. Previous studies have shown that AS events can better predict prognosis and guide treatment by reflecting modifications in the TME, including in gastric cancer (22), colorectal cancer (23), and glioblastomas (24). However, few studies have focused on the effect of AS on the TME in LUAD.
The purpose of this study was to explore the prognostic value of AS by integrated bioinformatics analysis and to understand the potential relationship between AS and the TME characteristics. Firstly, a novel prognostic signature based on the prognostic AS events was constructed and further validated by Kaplan-Meier (K-M) survival curves and receiver operating characteristic (ROC) curves. Furthermore, the correlation between this prognostic signature and the TME was assessed through several different algorithms. Finally, a potential SFs-AS regulatory network was established to explore the regulatory mechanisms of AS events in LUAD. This study may contribute to the accurate prognosis of LUAD and provide insights into the development of novel therapeutic targets. We present the following article in accordance with the TRIPOD reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1531/rc).
Methods
Data collection and processing
The research design and procedures were overviewed in Figure 1. The gene expression profiles and survival data of patients with LUAD (n=486) were extracted from TCGA database (https://www.cancer.gov/). The AS events and their percent-splice-in (PSI) values of patients with LUAD (n=513) were obtained from the TCGA SpliceSeq database (https://bioinformatics.mdanderson.org/TCGASpliceSeq) (25), and the PSI values (ranging from 0 to 1) were applied to quantify the likelihood of each AS events (26). To ensure the accuracy of the AS events, a portion of samples with PSI values <75% was excluded from further study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Identification of prognostic AS events and construction of a prognostic signature
Prognostic AS events were identified by univariate Cox regression analyses, and AS events with P value <0.001 were incorporated for further studies. AS events were visualized by Upset plots using the ‘UpSet’ R package (v1.3.3). Volcano and bubble plots were graphed to show each type of prognostic AS events. Least Absolute Shrinkage and Selection Operator (LASSO) regression was performed to screen the robust prognostic AS events using the ‘glmnet’ R package (v3.0-2), and these AS events were eligible for inclusion in subsequent multivariate Cox regression model. Finally, the following AS prognostic signature was constructed: , where β represents the regression coefficient of each AS event.
Evaluation of prognostic signature
To evaluate the accuracy of the survival analysis, 22 patients with incomplete survival or clinical data were excluded and finally, 464 patients with LUAD were included. These 464 LUAD patients were classified into high- and low-risk groups using the median risk score as a cutoff. Kaplan-Meier survival curves were plotted with ggsurvplot from the ‘survminer’ R package (v0.4.6). The time-dependent ROC curve was constructed and the area under the curve (AUC) value was calculated using the ‘timeROC’ R package (v0.4) to assess the prediction accuracy of the prognostic signature.
Identification and analysis of the TME
The Estimation of Stromal and Immune cells in Malignant Tumor tissues using Expression data (ESTIMATE) algorithm (27) was applied to analyze the overall stromal and immune components and tumor purity for each LUAD sample using ‘ESTIMATE’ R package (v1.1.0, https://bioinformatics.mdanderson.org/estimate/rpackage.html). The immune score or stromal score was calculated to represent the amount of overall immune or stromal components in the TME. The ESTIMATE score is the sum of the immune score and the stromal score, and represents the comprehensive proportion of both components in the TME. Tumor purity was defined as the percentage of malignant cells in a solid tumor sample. Differences in stromal, immune, and ESTIMATE scores and tumor purity between the two groups were visualized using violin plots.
The Microenvironment Cell Population (MCP)-counter algorithm (28) was applied to determine the absolute abundance of 8 immune cell types [including T cells, CD8 cells, natural killer (NK) cells, myeloid dendritic cells, cytotoxic lymphocytes, B lineage, monocytic lineage, and neutrophils], endothelial cells, and fibroblasts in LUAD tissues from transcriptional data. Differences in the abundance of cell populations between the two groups were visualized using boxplots. Additionally, the correlation between cell abundance and risk score was determined by Spearman rank correlation coefficient.
The single-sample Gene-Set Enrichment Analysis (ssGSEA) algorithm (29) was applied to calculate the immune signature score, implemented in the ‘GSVA’ R package (v1.34.0, https://bioconductor.org/packages/release/bioc/html/GSVA.html). As an extension of Gene-Set Enrichment Analysis (GSEA), ssGSEA evaluates the enrichment score of a specific gene set in every single sample (30). A total of 29 immune signature-specific gene sets derived from a published report (31) were included in this study, and the differences in the immune signature scores between the two groups were visualized using boxplots.
Establishing the SFs-AS regulatory network
To explore the regulatory mechanisms of AS events, a SFs-AS regulatory network was constructed. The SFs data was obtained from the SpliceAid2 database (http://www.introni.it/splicing.html), including 404 SFs (available online: https://cdn.amegroups.cn/static/public/atm-22-1531-1.xlsx) from a previous study (32). The SF gene expression profile of LUAD patients was extracted from TCGA RNA-seq data. Pearson correlation analysis was applied to assess the correlation of SFs and PSI values of prognostic AS events with correlation coefficient >0.5 and P value <0.001, and the regulatory network was generated in Cytoscape (v3.8.0).
Statistical analysis
All statistical analyses were performed via R software (version 4.1.1, www.r-project.org). Differences in classified data were examined by Chi-square or Fisher’s exact test, and differences in continuous data were assessed by Mann-Whitney U test between high-risk group and low-risk group. Survival analysis was performed using log-rank tests. In all hypothesis tests, a two-sided P value <0.05 was considered statistically significant. Univariate and multivariate Cox regression analyses were performed using the ‘survival’ R package (v2.41.3).
Results
Basic clinical characteristics and integrated AS events in lung adenocarcinoma
The profile of integrated AS events for 572 LUAD patients was extracted from TCGA SpliceSeq database, including 513 LUAD tumor samples and 59 corresponding normal samples. The gene expression profiles and the clinical phenotype data was obtained from TCGA database for 484 LUAD patients, excluding 22 patients with incomplete survival data. Finally, a total of 464 patients with complete splicing sequence, RNA sequence, and survival data were included in this study. The detailed information for this cohort was summarized in website: https://cdn.amegroups.cn/static/public/atm-22-1531-2.xlsx.
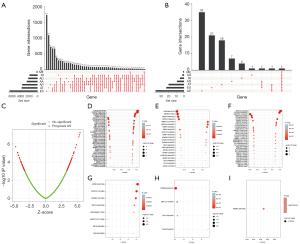
The integrated AS events profile was analyzed for all LUAD patients, and a total of 20,733 AS events from 10,005 genes were identified under the stringent filtering criteria. These AS events were classified into seven different splicing patterns: alternate acceptor site (AA), alternate donor site (AD), alternate promoter (AP), alternate terminator (AT), exon skip (ES), mutually exclusive exons (ME), and retained intron (RI). The specific numbers and intersections of different types of AS events in LUAD was visualized in an Upset plot diagram (Figure 2A). Among all AS events, ES (n=6,619) was the most commonly occurring splicing pattern, and ME (n=214) was the least frequent. Notably, each type of AS events may occur multiple times within a single gene, and a single gene may have more than one type of splicing pattern. A total of 5,684 genes containing 2 or more types of AS events were detected in the LUAD samples.
Identification of prognostic AS events in LUAD
A total of 127 AS events (available online: https://cdn.amegroups.cn/static/public/atm-22-1531-3.xlsx) in 89 genes were confirmed to be survival-associated in the LUAD samples (P<0.001), and the intersections among these prognostic AS events was visualized in an Upset plot diagram (Figure 2B). AP, AT, and ES events accounted for almost 90% of all AS events, and no ME events were survival associated. In addition, several genes had multiple types of AS events, for example, AP and AT splicing patterns in FAM72A were all associated with clinical outcomes in LUAD patients. The AS events are summarized in a volcano plot (Figure 2C). The prognostic AS events from each subtype are displayed in Figure 2D-2I.
Construction and validation of a prognostic signature for LUAD
LASSO regression (Figure 3A,3B) was performed to filter variables and avoid overfitting of the signature. The following 20 AS events (Table 1) were adopted for a multivariate Cox regression analysis to constructed a prognostic signature, including BEST3|23330|AT, HNRNPLL|53258|AT, TTC39C|44852|AP, CA5B|98313|ES, MEGF6|315|ES, SNAPC5|31278|ES, MECOM|67568|AP, FNBP1|87886|ES, ABCC6|34219|AT, ZNF707|85481|AD, ZNF580|52121|AP, ETV1|78828|AP, UBAP1|86151|ES, PLEC|85511|AP, ZNF268|25345|ES, FAXC|99934|ES, APOB|52793|AT, PSMF1|58475|AA, ANAPC15|17570|AD, and POLM|79445|RI. The risk score of each LUAD patient was calculated based on the prognostic signature, and all patients were stratified into a low- or high-risk group according to the cutoff value of the median risk score.
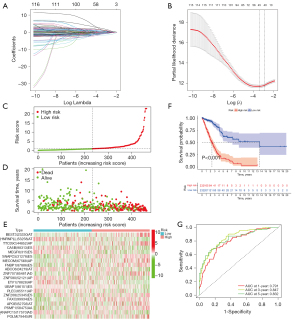
Table 1
| AS events | coef | HR | HR.95L | HR.95H | P value |
|---|---|---|---|---|---|
| BEST3|23330|AT | 1.2367 | 3.4442 | 1.1179 | 10.6119 | 0.0312 |
| HNRNPLL|53258|AT | −2.7518 | 0.0638 | 0.0063 | 0.6489 | 0.0200 |
| TTC39C|44852|AP | 0.8719 | 2.3915 | 0.9366 | 6.1064 | 0.0683 |
| CA5B|98313|ES | −0.7723 | 0.4620 | 0.1620 | 1.3169 | 0.1485 |
| MEGF6|315|ES | −1.2468 | 0.2874 | 0.1038 | 0.7957 | 0.0164 |
| SNAPC5|31278|ES | 3.5499 | 34.8110 | 1.5719 | 770.9016 | 0.0247 |
| MECOM|67568|AP | 1.8089 | 6.1036 | 1.6497 | 22.5826 | 0.0067 |
| FNBP1|87886|ES | 0.7399 | 2.0958 | 0.8365 | 5.2505 | 0.1143 |
| ABCC6|34219|AT | −1.8010 | 0.1651 | 0.0514 | 0.5302 | 0.0025 |
| ZNF707|85481|AD | 1.2666 | 3.5487 | 1.1690 | 10.7730 | 0.0254 |
| ZNF580|52121|AP | −0.9199 | 0.3986 | 0.1768 | 0.8985 | 0.0266 |
| ETV1|78828|AP | −2.0378 | 0.1303 | 0.0253 | 0.6712 | 0.0148 |
| UBAP1|86151|ES | −2.0564 | 0.1279 | 0.0298 | 0.5493 | 0.0057 |
| PLEC|85511|AP | −1.2916 | 0.2748 | 0.0543 | 1.3910 | 0.1185 |
| ZNF268|25345|ES | 1.9273 | 6.8709 | 1.7131 | 27.5567 | 0.0065 |
| FAXC|99934|ES | 2.8919 | 18.0274 | 4.1918 | 77.5282 | 0.0001 |
| APOB|52793|AT | 0.8648 | 2.3744 | 1.3553 | 4.1600 | 0.0025 |
| PSMF1|58475|AA | 4.5003 | 90.0445 | 1.5348 | 5282.6647 | 0.0303 |
| ANAPC15|17570|AD | −1.6290 | 0.1961 | 0.0751 | 0.5122 | 0.0009 |
| POLM|79445|RI | 1.8617 | 6.4348 | 1.7997 | 23.0082 | 0.0042 |
AS, alternative splicing; coef, regression coefficient; HR, hazard ratios.
The risk curve (Figure 3C) and scatter plot of the survival state (Figure 3D) suggested that patients with higher risk scores tended to have shorter overall survival. The differences in PSI values of the above AS events between the two groups was depicted in Figure 3E. The Kaplan-Meier curve (Figure 3F) illustrated that survival probability was significantly lower in the high-risk group compared to the low-risk group (P<0.001). The ROC curve (Figure 3G) demonstrated that the prognostic signature had a satisfactory prediction accuracy of LUAD patient prognosis, with an AUC of 0.791 at 1 year, 0.847 at 3 years, and 0.832 at 5 years, respectively.
Correlation between risk score and the TME
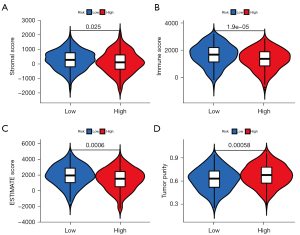
To explore whether the prognostic signature can reflect modifications of the TME in LUAD samples, correlation analyses between risk score and stromal/immune/ESTIMATE score, abundance of tumor-infiltrating immune cells (MCP-counter algorithm), and level of major immune signatures (ssGSEA algorithm) were performed. The ESTIMATE algorithm revealed that high-risk patients had significantly lower stromal, immune, and ESTIMATE scores (Figure 4A-4C). In addition, significantly higher tumor purity was observed in the high-risk group (Figure 4D).
The MCP-counter algorithm results indicated that high-risk patients exhibited a significantly lower abundance of T cells, CD8+ T cells, B lineage, NK cells, myeloid dendritic cells, neutrophils, and endothelial cells. However, a higher abundance of fibroblasts was observed in the high-risk group (Figure 5A). Additionally, Spearman correlation analysis showed that the prognostic signature exhibited stable negative correlation with the abundance of T cells (R=−0.22), CD8+ T cells (R=−0.14), B lineage (R=−0.099), myeloid dendritic cells (R=−0.21), neutrophils (R=−0.21), and endothelial cells (R=−0.017; Figure 5B-5G). Additionally, the abundance of fibroblasts was positively correlated with risk scores (R=0.15; Figure 5H). There was no significant correlation between risk score and other cell populations (P>0.05).
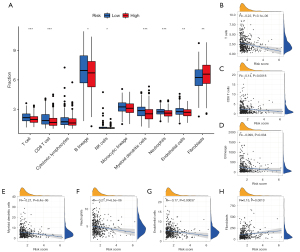
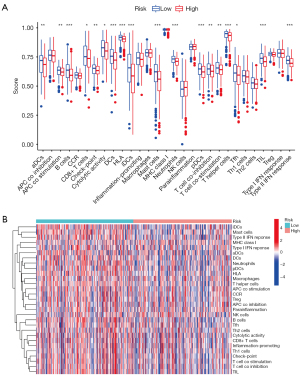
The differences in the level of 29 immune-related signatures were compared between the 2 groups. As shown in Figure 6A, high-risk patients exhibited significantly lower levels of majority immune-related signatures, including mast cells, neutrophils, antigen-presenting cells (APC) co-stimulation, dendritic cells (DCs), immature DCs (iDCs), activated DCs (aDCs), plasmacytoid DCs (pDCs), tumor infiltrating lymphocyte (TIL), B cells, CD8+ T cells, T helper cells, T follicular helper (Tfh) cells, T cell co-inhibition, T cell co-stimulation, cytolytic activity, human leucocyte antigen (HLA) type II interferon (IFN-II) response, and immune checkpoint expression. However, there were no significant differences in the levels of tumor-promoting inflammation, para-inflammation, APC co-stimulation, macrophages, NK cells, T helper 1 (Th1), Th2, Treg, CCR, major histocompatibility complex (MHC) class I, nor IFN-I response (P>0.05). The levels of 29 immune-related signatures of each patient are displayed in a heat map (Figure 6B). The above results indicated that the prognostic signature can also act as an important indicator of the TME.
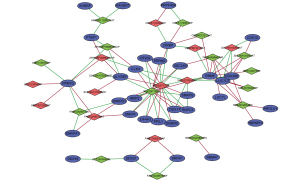
Establishing the SFs-AS regulatory network
To explore the potential regulatory mechanism between SFs and prognostic AS events in LUAD, a regulatory network (Figure 7) was established based on the results of the Pearson correlation tests. A total of 32 SFs (blue dots) were identified which were significantly related to 27 prognostic AS events, including 11 high-risk AS events and 16 low-risk AS events (available online: https://cdn.amegroups.cn/static/public/atm-22-1531-4.xlsx). Among them, several AS events were associated with multiple SFs, such as NEK2|9717|AT, NEK2|9718|AT, and SYNJ2|78244|AP. Meanwhile, several SFs were associated with multiple AS events, such as PPM1G and RBM5.
Discussion
In this study, we constructed a novel prognostic signature of LUAD based on 20 AS events and the robust performance was verified using K-M survival and ROC curve analyses. Furthermore, the correlation between this signature and the TME was assessed using several algorithms. Finally, a potential regulatory mechanism of AS events was explored.
Advances in next-generation sequencing technologies have gradually facilitated an understanding of the role of AS events in the development of cancers (33,34). Such roles include proliferation, invasion, and metastasis of tumor cells, as well as modification of the TME (35). The potential to use cancer-specific AS events as biomarkers for prognosis and treatment targets is gaining recognition (36,37). Recent studies have focused on exploring the prognostic value of AS events in malignancies such as colorectal cancer (38), breast cancer (39), hepatocellular carcinoma (40), and kidney cancer (41). LUAD is an aggressive tumor with increasing incidence and poor outcomes (42), and thus, the identification of biomarkers for diagnosis and therapy is urgently required. Li et al. recently identified prognostic predictors based on AS events with high performances for risk stratification in NSCLC patients (5). Cai et al. also established an AS-based prognostic model in LUAD patients (43). To further determine the ability of AS events to predict the prognosis of LUAD, we constructed a novel AS-based prognostic signature with a robust performance.
The TME plays a critical role in regulating tumor initiation and progression (44), which primarily consists of infiltrating stromal and immune cells. AS events may affect the outcomes of tumors by altering the TME, and few studies have focused on the effect of AS on the TME of LUAD. The current research further explored the differences in the TME among patients with different AS risk scores using several methods. First, we demonstrated that high-risk patients had lower stromal scores and immune scores, as well as higher tumor purity. Ma et al. found that LUAD patients with high stromal, immune, and ESTIMATE scores had better overall survival (P=0.066, P=0.0077, and P=0.0035, respectively) (45). Qu et al. showed a positive association between prognosis in LUAD patients and the immune and stromal scores (46). These findings indicated that poor prognosis of high-risk patients is closely related to the lower levels of infiltrating stromal and immune cells.
Our MCP-counter results revealed that the abundances of the majority immune cell types were significantly deficient in the high-risk group, including T cells, CD8 T cells, and B lineage cells that play a critical protective role in the adaptive immune response, as well as NK cells, myeloid dendritic cells, and neutrophils that play a vital role in the innate immune response. These findings indicated that the anti-tumor-related innate immunity and adaptive immune system of patients in the high-risk group were significantly suppressed. Apart from immune cells, stromal cells such as cancer-associated fibroblasts and endothelial cells, also play essential roles in regulating tumor progression (47). In this study, high-risk patients had a higher abundance of fibroblasts, which potentially related to a poor prognosis. Contrary to what we expected, the abundance of endothelial cells was lower in high-risk patients. This may be related to a higher proportion of functional tumor endothelial cells (TECs) (48).
The ssGSEA method was used to evaluate 29 major immune signatures in LUAD samples. Previous studies have shown that neutrophils and MCs are key players in innate immune responses (49,50), while DCs play a critical role in the crosstalk between innate and adaptive immunity through antigen processing and presentation (51). Low levels of neutrophils, DCs, MCs, and APC co-stimulation signature suggested that high-risk patients have not only impaired innate immune system but also weaken antigen presentation. Furthermore, B cells (52), Tfh cells (53), CD8+ T cells (54), Th cells (55), and IFN-II response (56) are major contributors to adaptive immune system, and low levels of these signatures suggested a severely impaired anti-tumor adaptive immunity in the high-risk group. Moreover, low levels of total TILs and cytolytic activity in the high-risk group further indicated the weaken anti-tumor effect, resulting in a poor prognosis of LUAD patients. In brief, high-risk patients are more likely to be accompanied by an inactive tumor immune microenvironment, which may contribute to poor prognosis.
There were some limitations to this research. First, the AS-based prognosis signature requires an additional independent patient cohort for external validation. Second, our research was based entirely on large-scale bioinformatics analysis, and further experimental validation is warranted.
In summary, we constructed an AS-based prognostic signature, using data from TCGA databases, which could effectively predict the survival of LUAD patients. Furthermore, we demonstrated that AS events could reflect modifications of the TME, which may be a crucial mechanism affecting the prognosis of LUAD patients. This work provided novel insights for the development of biomarkers and therapeutic targets in LUAD patients.
Acknowledgments
The authors would like to acknowledge the TCGA database for providing their platforms and those contributors for uploading their valuable datasets.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1531/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1531/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Ettinger DS, Akerley W, Bepler G, et al. Non-small cell lung cancer. J Natl Compr Canc Netw 2010;8:740-801. [Crossref] [PubMed]
- Duma N, Santana-Davila R, Molina JR. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin Proc 2019;94:1623-40. [Crossref] [PubMed]
- Li Y, Sun N, Lu Z, et al. Prognostic alternative mRNA splicing signature in non-small cell lung cancer. Cancer Lett 2017;393:40-51. [Crossref] [PubMed]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010;463:457-63. [Crossref] [PubMed]
- Pan Q, Shai O, Lee LJ, et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 2008;40:1413-5. [Crossref] [PubMed]
- Koh CM, Bezzi M, Low DH, et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 2015;523:96-100. [Crossref] [PubMed]
- Salton M, Kasprzak WK, Voss T, et al. Inhibition of vemurafenib-resistant melanoma by interference with pre-mRNA splicing. Nat Commun 2015;6:7103. [Crossref] [PubMed]
- Oltean S, Bates DO. Hallmarks of alternative splicing in cancer. Oncogene 2014;33:5311-8. [Crossref] [PubMed]
- David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev 2010;24:2343-64. [Crossref] [PubMed]
- Poulikakos PI, Persaud Y, Janakiraman M, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 2011;480:387-90. [Crossref] [PubMed]
- Avery-Kiejda KA, Zhang XD, Adams LJ, et al. Small molecular weight variants of p53 are expressed in human melanoma cells and are induced by the DNA-damaging agent cisplatin. Clin Cancer Res 2008;14:1659-68. [Crossref] [PubMed]
- Li ZX, Zheng ZQ, Wei ZH, et al. Comprehensive characterization of the alternative splicing landscape in head and neck squamous cell carcinoma reveals novel events associated with tumorigenesis and the immune microenvironment. Theranostics 2019;9:7648-65. [Crossref] [PubMed]
- Qi F, Li Y, Yang X, et al. Significance of alternative splicing in cancer cells. Chin Med J (Engl) 2020;133:221-8. [Crossref] [PubMed]
- Pietras K, Ostman A. Hallmarks of cancer: interactions with the tumor stroma. Exp Cell Res 2010;316:1324-31. [Crossref] [PubMed]
- Fridman WH, Dieu-Nosjean MC, Pagès F, et al. The immune microenvironment of human tumors: general significance and clinical impact. Cancer Microenviron 2013;6:117-22. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 2018;24:541-50. [Crossref] [PubMed]
- Choi H, Na KJ. Integrative analysis of imaging and transcriptomic data of the immune landscape associated with tumor metabolism in lung adenocarcinoma: Clinical and prognostic implications. Theranostics 2018;8:1956-65. [Crossref] [PubMed]
- Giraldo NA, Becht E, Vano Y, et al. Tumor-Infiltrating and Peripheral Blood T-cell Immunophenotypes Predict Early Relapse in Localized Clear Cell Renal Cell Carcinoma. Clin Cancer Res 2017;23:4416-28. [Crossref] [PubMed]
- Kim EK, Yoon SO, Jung WY, et al. Implications of NOVA1 suppression within the microenvironment of gastric cancer: association with immune cell dysregulation. Gastric Cancer 2017;20:438-47. [Crossref] [PubMed]
- Wan L, Yu W, Shen E, et al. SRSF6-regulated alternative splicing that promotes tumour progression offers a therapy target for colorectal cancer. Gut 2019;68:118-29. [Crossref] [PubMed]
- Razavi SM, Lee KE, Jin BE, et al. Immune Evasion Strategies of Glioblastoma. Front Surg 2016;3:11. [Crossref] [PubMed]
- Ryan MC, Cleland J, Kim R, et al. SpliceSeq: a resource for analysis and visualization of RNA-Seq data on alternative splicing and its functional impacts. Bioinformatics 2012;28:2385-7. [Crossref] [PubMed]
- Schafer S, Miao K, Benson CC, et al. Alternative Splicing Signatures in RNA-seq Data: Percent Spliced in (PSI). Curr Protoc Hum Genet 2015;87:11.16.1-11.16.14.
- Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [Crossref] [PubMed]
- Becht E, Giraldo NA, Lacroix L, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol 2016;17:218. [Crossref] [PubMed]
- Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009;462:108-12. [Crossref] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782-95. [Crossref] [PubMed]
- Seiler M, Peng S, Agrawal AA, et al. Somatic Mutational Landscape of Splicing Factor Genes and Their Functional Consequences across 33 Cancer Types. Cell Rep 2018;23:282-296.e4. [Crossref] [PubMed]
- Zhu S, Chen W, Wang J, et al. SAM68 promotes tumorigenesis in lung adenocarcinoma by regulating metabolic conversion via PKM alternative splicing. Theranostics 2021;11:3359-75. [Crossref] [PubMed]
- Qu S, Jiao Z, Lu G, et al. PD-L1 lncRNA splice isoform promotes lung adenocarcinoma progression via enhancing c-Myc activity. Genome Biol 2021;22:104. [Crossref] [PubMed]
- Climente-González H, Porta-Pardo E, Godzik A, et al. The Functional Impact of Alternative Splicing in Cancer. Cell Rep 2017;20:2215-26. [Crossref] [PubMed]
- Urbanski LM, Leclair N, Anczuków O. Alternative-splicing defects in cancer: Splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip Rev RNA 2018;9:e1476. [Crossref] [PubMed]
- Bonnal SC, López-Oreja I, Valcárcel J. Roles and mechanisms of alternative splicing in cancer - implications for care. Nat Rev Clin Oncol 2020;17:457-74. [Crossref] [PubMed]
- Shi JY, Bi YY, Yu BF, et al. Alternative Splicing Events in Tumor Immune Infiltration in Colorectal Cancer. Front Oncol 2021;11:583547. [Crossref] [PubMed]
- Deng Y, Zhao H, Ye L, et al. Correlations Between the Characteristics of Alternative Splicing Events, Prognosis, and the Immune Microenvironment in Breast Cancer. Front Genet 2021;12:686298. [Crossref] [PubMed]
- Wu F, Chen Q, Liu C, et al. Profiles of prognostic alternative splicing signature in hepatocellular carcinoma. Cancer Med 2020;9:2171-80. [Crossref] [PubMed]
- Wang Y, Chen SX, Rao X, et al. Modulator-Dependent RBPs Changes Alternative Splicing Outcomes in Kidney Cancer. Front Genet 2020;11:265. [Crossref] [PubMed]
- Chen H, Carrot-Zhang J, Zhao Y, et al. Genomic and immune profiling of pre-invasive lung adenocarcinoma. Nat Commun 2019;10:5472. [Crossref] [PubMed]
- Cai Q, He B, Zhang P, et al. Exploration of predictive and prognostic alternative splicing signatures in lung adenocarcinoma using machine learning methods. J Transl Med 2020;18:463. [Crossref] [PubMed]
- Quail DF, Joyce JA. The Microenvironmental Landscape of Brain Tumors. Cancer Cell 2017;31:326-41. [Crossref] [PubMed]
- Ma Q, Chen Y, Xiao F, et al. A signature of estimate-stromal-immune score-based genes associated with the prognosis of lung adenocarcinoma. Transl Lung Cancer Res 2021;10:1484-500. [Crossref] [PubMed]
- Qu Y, Cheng B, Shao N, et al. Prognostic value of immune-related genes in the tumor microenvironment of lung adenocarcinoma and lung squamous cell carcinoma. Aging (Albany NY) 2020;12:4757-77. [Crossref] [PubMed]
- Pereira BA, Vennin C, Papanicolaou M, et al. CAF Subpopulations: A New Reservoir of Stromal Targets in Pancreatic Cancer. Trends Cancer 2019;5:724-41. [Crossref] [PubMed]
- Yang D, Guo P, He T, et al. Role of endothelial cells in tumor microenvironment. Clin Transl Med 2021;11:e450. [Crossref] [PubMed]
- Eruslanov EB, Bhojnagarwala PS, Quatromoni JG, et al. Tumor-associated neutrophils stimulate T cell responses in early-stage human lung cancer. J Clin Invest 2014;124:5466-80. [Crossref] [PubMed]
- Dudeck A, Köberle M, Goldmann O, et al. Mast cells as protectors of health. J Allergy Clin Immunol 2019;144:S4-S18. [Crossref] [PubMed]
- Gardner A, Ruffell B. Dendritic Cells and Cancer Immunity. Trends Immunol 2016;37:855-65. [Crossref] [PubMed]
- Kim SS, Sumner WA, Miyauchi S, et al. Role of B Cells in Responses to Checkpoint Blockade Immunotherapy and Overall Survival of Cancer Patients. Clin Cancer Res 2021;27:6075-82. [Crossref] [PubMed]
- Zhang X, Dai R, Li W, et al. Abnormalities of follicular helper T-cell number and function in Wiskott-Aldrich syndrome. Blood 2016;127:3180-91. [Crossref] [PubMed]
- Lu C, Yang D, Klement JD, et al. SUV39H1 Represses the Expression of Cytotoxic T-Lymphocyte Effector Genes to Promote Colon Tumor Immune Evasion. Cancer Immunol Res 2019;7:414-27. [Crossref] [PubMed]
- Gao S, Hsu TW, Li MO. Immunity beyond cancer cells: perspective from tumor tissue. Trends Cancer 2021;7:1010-9. [Crossref] [PubMed]
- Martinez-Sabadell A, Arenas EJ, Arribas J. IFNgamma Signaling in Natural and Therapy-Induced Antitumor Responses. Clin Cancer Res 2021; Epub ahead of print. [Crossref] [PubMed]
(English Language Editor: J. Teoh)


