
DNAJC12 activated by HNF1A enhances aerobic glycolysis and drug resistance in non-small cell lung cancer
Introduction
Lung cancer is one of the leading causes of cancer-related death worldwide (1). It is estimated that more than 7,333,000 patients are diagnosed with lung cancer every year and it causes approximately 6,102,000 deaths annually in China (2). Non-small cell lung cancer (NSCLC) is the most common type of lung cancer, accounting for about 80% of reported cases (3). Platinum, especially cisplatin (DDP)-based chemotherapy is a main treatment method for NSCLC after surgical resection (4). However, prolonged use of DDP often induces chemoresistance in tumor cells, allowing them escape apoptosis (5). Therefore, it is an urgent need to reveal the mechanism underlying DPP resistance in NSCLC.
Cancer cells are characterized by metabolic deregulation, including aerobic glycolysis, which is also known as the “Warburg effect” (6). Through increasing of the uptake and consumption of glucose, cancer cells gain enough energy to maintain their rapid growth, even in the presence of high oxygen concentration (7). Studies have shown that glycolysis is closely related to the prognosis of NSCLC patients. For instance, Smolle et al. (8) showed that the expression of GLUT1, the prime glucose transporter, was associated with decreased overall survival of patients with NSCLC. Monaco et al. (9) revealed that patients with total lesion glycolysis (TLG) values lower than the median values had improved overall survival compared to patients with higher TLG. Yao et al. (10) obtained 200 glycolysis-related genes from Gene Set Enrichment Analysis (GSEA) and found 46 genes were significantly associated with the overall survival of patients with NSCLC. Also, it has demonstrated that the aerobic glycolysis plays crucial roles in accelerating cancer cell growth, migration, and drug resistance (11,12). Targeting of aerobic glycolysis is an innovative idea to overcome drug resistance in cancers (13,14).
The gene DnaJ heat shock protein family member C12 (DNAJC12) belongs to the heat shock protein (HSP) family, which act as molecular chaperones (15,16). Recently, studies have demonstrated that DNAJC12 is overexpressed and implicated in carcinogenesis, including gastric cancer (17) and rectal cancer (18). Using bioinformatics technology, we found that DNAJC12 is upregulated in lung adenocarcinoma (LUAD). Consistently, Li et al. (19) recently reported that DNAJC12 was overexpressed in lung cancer, and overexpression of DNAJC12 significantly promoted lung cancer cell growth and migration by increasing βcatenin expression. Noticeably, accumulated evidence has verified that the β-catenin signaling is closely implicated in regulating the aerobic glycolysis and drug resistance in lung cancer (20-23), suggesting that DNAJC12 may participate in regulating the aerobic glycolysis and drug resistance of lung cancer through activating the β-catenin signaling. However, the role of DNAJC12 in modulating aerobic glycolysis and DDP resistance in NSCLC still remains unclear.
Hepatocyte nuclear factor 1-alpha (HNF1A) is an endoderm-restricted transcription factor and has been reported to induce long non-coding RNA (lncRNA) HCG18 expression which promoted gastric cancer progression (24). Also, HNF1A has been identified to serve as an oncogene in pancreatic cancer to increase cancer stem cell properties (25). Using bioinformatics technology, we found that HNF1A expression was positively correlated with DNAJC12, and HNF1A can bind to the promoter region of DNAJC12. Noticeably, it has been reported that overexpression of HNF1A causes significant enhancement in the drug resistance of colorectal cancer (26). Thus, we assumed that DNAJC12 transcription may be increased by HNF1A and then accelerates aerobic glycolysis and enhances drug resistance to DDP in NSCLC.
Although it has been demonstrated that DNAJC12 was overexpressed and served as an oncogene in lung cancer through regulating βcatenin signaling (19), DNAJC12 roles in modulating aerobic glycolysis and DDP resistance in NSCLC still remain unclear, as well as the underlying mechanisms. In this study, we explored the role of DNAJC12 in aerobic glycolysis and DDP resistance of NSCLC, and studied whether DNAJC12 transcription could be modulated by HNF1A for the first time using both in vitro (cell lines) and in vivo assays through construction of the xenotransplantation model in 6-week-old BALB/c nude mice. We present the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1475/rc).
Methods
Tissue samples
A total of 60 NSCLC tissues and normal tissues adjacent to the cancer tissues were obtained from patients with primary NSCLC at the Department of Thoracic Surgery, Shanghai General Hospital between February, 2013 and August, 2015. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of Shanghai General Hospital (No. 2018KY086) and informed consent was taken from all the patients.
Bioinformatics analysis
We used the University of Alabama Cancer (UALCAN; http://ualcan.path.uab.edu/analysis.html) and Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/) databases to analyze the expression pattern of DNAJC12 in NSCLC tissues. The starBase database (https://starbase.sysu.edu.cn/) was applied to analyze the expression levels of HNF1A in NSCLC samples, and the correlation between the expression levels of DNAJC12 and HNF1A in NSCLC. The JASPAR (https://jaspar.genereg.net/) database was used to predict the binding sites of transcription factor HNF1A in the DNAJC12 promoter region.
Cell culture and DDP-resistance cell line construction
A human lung epithelial cell line BEAS-2B and 4 human lung cancer cell lines A549, NCI-H2106, NCI-H1975, and NCI-H1650 were purchased from American Type Culture Collection (ATCC; Manassas, VA, USA). Then, NCI-H1975/DDP and NCI-H1650/DDP, 2 cisplatin-resistant cell lines were established by explosion to a series of stepwise‑increased concentrations of DDP. The A549 cells were maintained in F-12K medium, while NCI-H2106, NCI-H1975, NCI-H1650, NCH-H1975/DDP, and NCI-H1650/DDP cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium, with 10% fetal bovine serum (FBS). To maintain the resistance, 1 µg/mL DDP was added to cell cultures of NCH-H1975/DDP and NCI-H1650/DDP cells. All cells were maintained in a humid cell incubator with 5% CO2 at 37 ℃. Cell culture medium and FBS were obtained from Thermo Fisher Scientific, Inc. (Waltham, MA, USA). The NCH-H1975/DDP and NCI-H1650/DDP cells were administrated with 10 µM cisplatin (DDP, Sigma Aldrich, St. Louis, MO, USA) for 24 hours.
Cell transfection
The overexpressing plasmid used to upregulate DNAJC12 (OE-DNAJC12) and HNF1A (OE-HNF1A) expression in NSCLC cells were purchased from Origene (cat. RC201931 and RC211201; Beijing, China). The cells were plated into 6-well plates and transfected with 2 µg of OE-DNAJC12 or OE-HNF1A with the help of Lipofectamine 3000 (Invitrogen, Thermo Fisher Scientific) when cell confluence reached about 50%. The lentivirus short hairpin RNA (shRNA) vectors used to downregulate DNAJC12 (sh-DNAJC12), HNF1A (sh-HNF1A) and β-catenin (sh-β-catenin) were synthesized by GenePharma (Shanghai, China) and introduced into cells using polybrene (5 µg/mL). The infected cells were cultured in RPMI-1640 medium with puromycin (4 µg/mL) for 14 days to establish stable cell lines to be used in the animal assay.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
TRIzol reagent (Invitrogen, USA) was used for the total RNA extraction. After that, the complementary DNA (cDNA) was synthesized using PrimeScript RT Master Mix kit (RR036A; Takara, Shiga, Japan). Then, the PCR detection was carried out with 2× SYBR Green PCR Mastermix (Solarbio, Beijing, China) in 7500 Real-Time PCR System (Applied Biosystems, Forster City, CA, USA) with the following reactions: 95 ℃ for 1 minute, 39 cycles of 95 ℃ for 15 seconds and 60 ℃ for 1 minute. The relative expressions of messenger RNA (mRNA) were calculated in the light of the 2−∆∆Cq method (27). The primers are listed in Table 1.
Table 1
| Gene | Forward (5'-3') | Reverse (5'-3') |
|---|---|---|
| DNAJC12 | CGAAGCTCACTGTGCCTCTT | GGATGCTTGTCTGGGTGACA |
| HNF1A | ACAGCTTGGAGCAGACATCC | CTGCTTGGTGGGCGTGAG |
| β-catenin | CTGAGGAGCAGCTTCAGTCC | CCATCAAATCAGCTTGAGTAGCC |
| β-actin | ACAGAGCCTCGCCTTTGCC | TTCTCCATGTCGTCCCAGTTG |
DNAJC12, DnaJ heat shock protein family member C12; HNF1A, hepatocyte nuclear factor 1-alpha.
Western blotting
Lysis buffer (Solarbio, Beijing, China) with 1% protease inhibitor (Solarbio) were used for total protein isolation. After centrifugation at 4 ℃ for 30 minutes, protein concentrations was tested using bicinchoninic acid (BCA) Protein Assay kits (Thermo Fisher Scientific). Then, the proteins were loaded to 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel and separated by electrophoresis. Then, transformation to polyvinylidene difluoride membranes (PVDF; Millipore, Billerica, MA, USA) was performed. After that, the membranes were blocked in 5% non-fat milk for 1 hour at room temperature and immersed in primary antibodies overnight at 4 ℃ [anti-β-actin antibody (1:5,000 dilution; cat. ab8226, Abcam, Cambridge, MA, USA) and anti-DNAJC12 antibody (1:2,000 dilution; cat. ab167425, Abcam)]. The membranes were then incubated with the horseradish peroxidase (HRP)-conjugated secondary antibodies at room temperature for 1 hour. Protein signaling was measured by using the ProfiBlot-48 (Tecan, Zurich, Switzerland) with the help of electrochemiluminescence (ECL) reagent (Millipore, USA) and quantified using Image J v2.1.4.7 (National Institutes of Health, Bethesda, MD, USA).
Double luciferase gene reporter measurement
Sequences of DNAJC12 promoter regions were inserted into the luciferase reporter pGL3 vectors. Then, the cells were co-transfected with the pGL3 vectors and OE-NC, OE-HNF1A, sh-NC, or sh-HNF1A. After 48 hours, cells in each group were collected for luciferase activity detection based on the Dual-Luciferase Reporter Assay protocol (Promega, Madison, WI, USA).
Detection of lactate production, glucose consumption, and adenosine triphosphate (ATP) levels
The lactate production, glucose consumption, and ATP levels were evaluated as previously reported (28). Cells were placed into 6-well plates at a density of 2×105/well and cultured at 37 ℃ for 48 hours, then the lactate production of cells was examined using Lactate Colorimetric Assay Kits (cat. no. K627, Biovision, Milpitas, CA, USA). Supernatant was collected from cells kept in FBS-free medium for hour, and used for the measurement of lactate production. The reaction mixture was incubated for 30 minutes at room temperature in the dark. The lactate levels were measured at 450 nm in a microplate reader.
Glucose consumption was examined using Glucose Uptake Colorimetric Assay Kit (cat. no. K676, Biovision, Milpitas, CA, USA). Briefly, cells with lentivirus infection were collected and placed into 96-well plates at a density of 1×104 cells/well. Then, the cells were incubated with Krebs-Ringer-Phosphate-HEPES (KRPH) buffer (100 µL) for 40 minutes, and 10 µL 2-deoxyglucose (10 mM) for 20 minutes, and Reaction Mix A for 1 hour at 37 ℃ to deprive glucose. Then, the cells were probed with 90 µL extraction buffer for 40 minutes at 90 ℃, followed by an ice bath for 5 minutes. Each well was then added reaction Mix B, and centrifugation at 12,000 rpm at 4 ℃ for 2 minutes was performed. The optical density (OD) value at 412 nm was measured using a microplate analyzer for the supernatant.
An ATP Colorimetric Assay Kit purchased from Sigma-Aldrich (cat. no. MAK1900) was used for ATP level detection based on the manufacturer’s protocols. Cells (5×105) were harvested and extracted in 100 µL of the ATP Assay Buffer. The cells were centrifuged at 12,000 rpm for 5 minutes and the supernatant was used for ATP measurement. The reaction mixture was incubated for 30 minutes at room temperature, protected from light, and measured at 570 nm in a microplate reader.
Cell Counting Kit-8 assay
The NSCLC cells at logarithmic growth phase were adjusted to a density of 2×104 cells/mL and placed into 96-well plates (100 µL per well). Then, 10 µL of CCK-8 (Abcam) was added and the cells were cultured at 37 ℃ for another 4 hours after cell transfection. The OD at 450 nm was measured on microplate reader (Tecan Infinite M200 Micro Plate Reader; LabX, Männedorf, Switzerland).
Flow cytometry assay
An FITC-Annexin V Apoptosis Detection Kit (Becton, Dickinson, and Co. Biosciences, San Diego, CA, USA) was used to determine cell apoptosis rates according to the manufacturer’s instructions. The cells were analyzed by a Beckman FC500 flow cytometer (Beckman Coulter, Inc., Brea, CA, USA). The data were analyzed by FlowJo 7.6 software (https://www.flowjo.com/).
Animal assay
Animal experiments were performed under a project license (No. 2018AWS0095) granted by the Ethics Committee of Shanghai General Hospital, in compliance with the institutional guidelines for the care and use of animals. A protocol was prepared before the study without registration. The 6-week-old BALB/c nude mice with specific-pathogen-free (SPF) grade were obtained from the Animal Center of Air Force Medical University (Shanghai, China). The mice were housed at 25 ℃ with 55% humidity in a 12 light/12 dark cycle with ad libitum access to water and food. The weight of the included mice were 20±2 g and the mice were fed in same condition to minimize potential confounders. A total of 10 mice were randomly divided into the sh-NC and sh-DNAJC12 group, with each group containing 5 mice. The NCI-H1650 cells with sh-NC or sh-DNAJC12 stable transfection were used to build the model. The mice armpits were injected with 1×106 cells following 7 days of accommodation. Tumor volume was measured every 3 days. At 28 days after injection, or when the diameter of tumors reached over 1 cm, mice were sacrificed by cervical dislocation. Mice were considered dead when their breathing and heartbeat stopped, they had no reflexes, and the body became cold. Then, the tumors were harvested and weighed, and the tissues were used for western blotting, qRT-PCR and hematoxylin and eosin staining.
In addition, hematoxylin and eosin staining was used to assess the morphological changes of the tumor tissues. In brief, tumor samples were washed, dehydrated in 70%, 80% and 90% ethanol solutions, immersed in equal volumes of absolute alcohol and xylene for 15 min, transparentized in xylene and immersed in paraffin and xylene for 15 min. Following being embedded in paraffin, the tissues were cut into 4 µm-thick sections, and the sections were dewaxed and hydrated, following by staining with hematoxylin solution for 3 min. The sections were then washed with water and immersed in eosin solution for 3 min. The staining was assessed under a light microscope (CKX41; Olympus Corporation).
Statistical analysis
The data from three independent assays were presented as the mean ± standard deviation (SD), and P<0.05 was considered statistically significant. The statistical analyses were carried out using the software SPSS 21.0 (IBM Corp., Armonk, NY, USA). Student’s t-test was used to analyze the difference between two groups. One-way analysis of variance (ANOVA) followed by Dunnett’s test was adopted to compare more than two groups. Pearson correlation coefficient was adopted to analyze the correlation between DNAJC12 expression and HNF1A expression in NSCLC tissues.
Results
DNAJC12 is overexpressed in NSCLC tissues and cells
First, we assessed the expression levels of DNAJC12 in NSCLC tissues and cells. The UALCAN database showed that DNAJC12 was among the top 25 upregulated genes in LUAD (Figure 1A), which was confirmed by the GEPIA database (Figure 1B). To verify this, we detected DNAJC12 expression in 60 paired NSCLC tissues and normal tissues using qRT-PCR assay. The results showed that the level of DNAJC12 was significantly higher in NSCLC tissues than in normal tissues (Figure 1C). We also detected DNAJC12 protein levels in 4 matched cancer tissues and normal tissues, randomly selected from the 60 paired tissues, using western blotting assay. Consistently, DNAJC12 level was elevated in cancer tissues as compared with the normal tissues (Figure 1D). In addition, DNAJC12 levels in NSCLC cell lines, including NCI-H1975, A549, NCI-H2106, and NCI-H1650 cells were obviously higher than that of BEAS-2B cells (Figure 1E,1F). These results demonstrated that DNAJC12 was overexpressed in NSCLC.
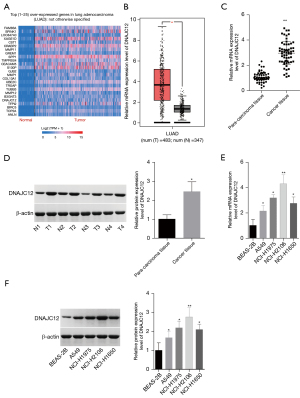
DNAJC12 accelerates aerobic glycolysis in NSCLC cells
Then, we explored the role of DNAJC12 in regulating aerobic glycolysis in NSCLC cells using the gain- and loss-of-function assays. As NCI-H1975 and NCI-H1650 cells showed medium expression levels of DNAJC12 among the 4 detected cell lines, NCI-H1975 and NCI-H1650 cells were chosen for the following studies. The level of DNAJC12 was significantly increased following cell transfection with OE-DNAJC12, while decreased when cells were infected with sh-DNAJC12 (Figure 2A,2B). Compared with the control group, DNAJC12 overexpression significantly increased ATP levels, lactate production, and glucose consumption, while silencing of DNAJC12 caused the opposite results (Figure 2C-2E). These results demonstrated that DNAJC12 promoted aerobic glycolysis in NSCLC cells.
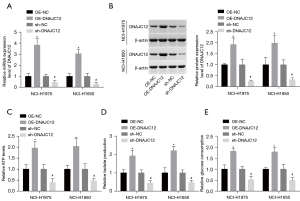
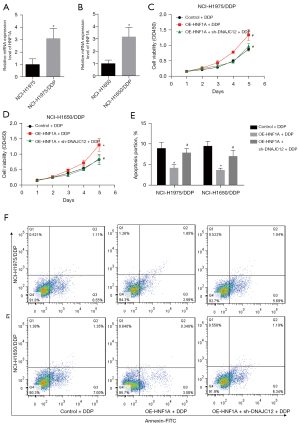
DNAJC12 promotes cell growth and enhances DDP resistance in NSCLC
We assessed the role of DNAJC12 in modulating cell growth and drug resistance. Cell growth was significantly enhanced when DNAJC12 was overexpressed in NCI-H1975 and HCI-H1650 cells (Figure 3A,3B), while cell apoptosis was reduced (Figure 3C,3D). However, downregulation of DNAJC12 inhibited cell growth and increased cell apoptosis (Figure 3A-3D). In addition, DNAJC12 levels in NCH-H1975/DDP and NCI-H1650/DDP cells were higher than those in their parental cells (Figure 3E-3H). Moreover, overexpression of DNAJC12 reversed the inhibition of cell growth induced by DDP and reduced cell apoptosis, while depletion of DNAJC12 enhanced the role of DDP in inhibiting cell growth (Figure 3I,3J) and inducing cell apoptosis (Figure 3K,3L). These results demonstrated that DNAJC12 promoted NSCLC cell growth and enhanced drug resistance to DDP.
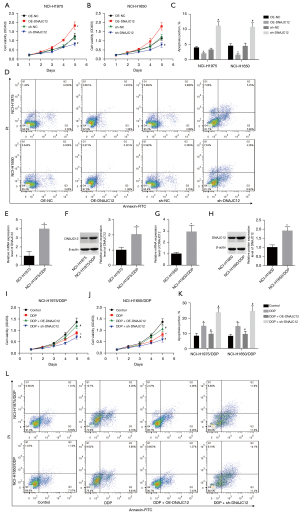
HNF1A promotes DNAJC12 expression
Through bioinformatics, we found that DNAJC12 expression was positively correlated with the expression level of HNF1A, which was also upregulated in lung cancer tissues (Figure 4A,4B). We then used qRT-PCR to detect HNF1A expression in 60 paired cancer tissues and normal tissues. The expression level of HNF1A was found to be higher in cancer tissues compared with the normal tissues (Figure 4C) and its expression showed a positive correlation with DNAJC12 expression in NSCLC tissues (Figure 4D). As HNF1A is a transcription factor, we then applied JASPAR databases to determine whether HNF1A may regulate DNAJC12. The DNA motif of HNF1A and 2 predicted binding sites of HNF1A in DNAJC12 promoter are illustrated in Figure 4E,4F. Luciferase gene reporter assay was then applied to verify these findings. The expression of HNF1A was significantly increased when the cells were transfected with OE-HNF1A, while decreased when cells were infected with sh-HNF1A (Figure 4G). Overexpression of HNF1A induced a significant increase in the luciferase activity and vice versa (Figure 4H), indicating that HNF1A positively modulated DNAJC12 expression in NSCLC cells. This was further confirmed by the qRT-PCR and western blot assays (Figure 4I,4J). These results illustrated that HNF1A was overexpressed in NSCLC and positively promoted DNAJC12 expression.
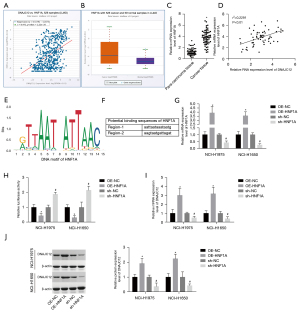
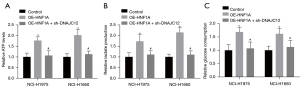
HNF1A promotes aerobic glycolysis and drug resistance through increasing DNAJC12 expression in NSCLC cells
Next, we explored whether HNF1A promoted aerobic glycolysis and drug resistance by increasing DNAJC12 expression in NSCLC cells in vitro. The ATP levels, lactate production, and glucose consumption were significantly increased when HNF1A expression was upregulated, whereas this tendency was abolished with silencing of DNAJC12 (Figure 5A-5C). In addition, HNF1A expression was elevated in NCH-H1975/DDP and NCI-H1650/DDP cells compared to their parental cells (Figure 6A,6B). Overexpression of HNF1A enhanced cell growth which had been inhibited by DDP (Figure 6C,6D) and reduced DDP-induced cell apoptosis (Figure 6E,6F), while depletion of DNAJC12 rescued this. These findings suggested that HNF1A facilitated aerobic glycolysis and drug resistance through increasing DNAJC12 expression in NSCLC cells.
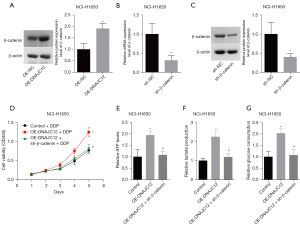
Silencing of β-catenin reverses DNAJC12-mediated cell growth and aerobic glycolysis
Li et al. (19) recently reported that DNAJC12 was overexpressed in lung cancer and significantly promoted lung cancer cell growth and migration by increasing βcatenin expression. And, evidence has verified that the β-catenin signaling is closely implicated in regulating the aerobic glycolysis and drug resistance in lung cancer (20-23). Thus, we conjectured β-catenin may play a role in DNAJC12-mediated cell growth and aerobic glycolysis. To this end, the rescue experiments were carried out. Upregulation of DNAJC12 significantly increased β-catenin expression in NCI-H1650 cells as compared with the control group (Figure 7A), while β-catenin expression were significantly decreased when the cells were infected with the sh-β-catenin (Figure 7B,7C). In addition, downregulation of β-catenin significantly inhibited cell growth mediated by DNAJC12 under DDP treatment (Figure 7D), and decreased the ATP level (Figure 7E), lactate production (Figure 7F) and glucose consumption (Figure 7G). These results revealed that DNAJC12 promoted cell growth and aerobic glycolysis in NSCLC through upregulating β-catenin expression.
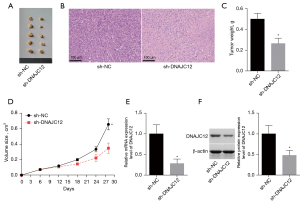
Downregulation of DNAJC12 inhibits tumor growth in vivo
We assessed the role of DNAJC12 in modulating the tumorigenesis of NSCLC cells. Figure 8A showed the tumor pictures of the sh-DNAJC12 and sh-NC group. The sh-NC group exhibited a relative uniform distribution of tumor cells and less necrotic areas, while large areas of necrosis were observed in the sh-DNAJC12 group (Figure 8B). Also, compared with the sh-NC group, both tumor weight and volume were decreased in the sh-DNAJC12 group (Figure 8C,8D), with a decrease expression level of DNAJC12 in tumor tissues (Figure 8E,8F). This result demonstrated that downregulation of DNAJC12 inhibited tumor growth in vivo.
Discussion
In the present study, we explored the role of DNAJC12, a member of the HSP family, in modulating cell aerobic glycolysis and drug resistance for the first time. Our results demonstrated that DNAJC12 overexpression promoted cell aerobic glycolysis with increased levels of ATP, lactate production, and glucose consumption in NSCLC cells. In addition, DNAJC12 expression was upregulated in DDP-resistant cells, and overexpression of DNAJC12 significantly enhanced cell resistance to DDP.
The role of DNAJC12 in several kinds of cancers has been gradually revealed. Uno et al. (17) found that DNAJC12 was overexpressed in gastric cancer tissues, which was closely linked to lymphatic involvement, lymph node metastasis, infiltrative growth type, and advanced stage and shorter overall survival. In addition, knockdown of DNAJC12 significantly repressed cell growth and invasiveness in gastric cancer. He et al. (18) identified that high expression of DNAJC12 served as a negative predictive factor for the response to neoadjuvant concurrent chemoradiotherapy (CCRT) and was strongly linked to shorter survival time in patients with rectal cancers receiving neoadjuvant CCRT and surgery. De Bessa et al. (29) reported that DNAJC12 mRNA expression was significantly associated with estrogen receptor-positive status in breast cancer, and 17β-estradiol treatment could increase DNAJC12 expression in breast cancer MCF-7 cells. It has also been reported recently that when DNAJC12 was overexpressed in lung cancer, knockdown of DNAJC12 inhibited the proliferation, colony formation, migration, invasion in vitro, and tumorigenesis in vivo of lung cancer cells through inhibiting the expression of β-catenin (19). Consistently, we found that DNAJC12 showed a higher expression pattern in NSCLC tissues and cells, and overexpression of DNAJC12 significantly promoted NSCLC cell growth, and inhibited apoptosis.
It has been verified that DNAJC12 is a positive regulator of β-catenin in lung cancer (19), and the β-catenin is closely implicated in regulating the aerobic glycolysis in lung cancer (20,21). These results indicate that DNAJC12 may participate in regulating the aerobic glycolysis of lung cancer through activating the β-catenin signaling. As respected, we found that DNAJC12 overexpression promoted aerobic glycolysis in NSCLC cells, which was rescued by β-catenin downregulation. Consistently, it has been demonstrated that other members of the HSP family are also implicated in aerobic glycolysis. For instance, Xu et al. (30) revealed that HSP90 promoted the glycolysis and proliferation of hepatocellular carcinoma cells in a PKM2 dependent manner. Also known as HSPD1, HSP60 was found to be upregulated in myeloma cells, and silencing of HSP60 inhibited glycolysis (31). The novel C-terminal HSP90 inhibitor KU757 could overcome the increase in aerobic glycolysis induced by lenvatinib treatment in lenvatinib-resistant follicular thyroid cancer cells (32).
In addition, hyper-activation of the β-catenin signaling plays an important role in the drug resistance of lung cancer (22,23), suggesting that DNAJC12 may participate in regulating the DDP resistance of lung cancer through activating the β-catenin signaling Moreover, evidence has demonstrated that aerobic glycolysis deregulation can help cancer cells to increase drug resistance. Targeting metabolic disorder is a promising method for cancer intervention (33,34). As DNAJC12 could promote aerobic glycolysis, we conjectured that DNAJC12 maybe also implicated in modulating cell drug resistance in NSCLC through regulating β-catenin signaling-mediated aerobic glycolysis. As expected, we observed that DNAJC12 expression was upregulated in DDP-resistant cells, and overexpression of DNAJC12 significantly increased cell growth and reduced cell apoptosis in the presence of DDP. Besides, we found that downregulation of β-catenin reversed DNAJC12-mediated enhancement of cell viability under DDP treatment, indicating that DNAJC12 accelerated cellular drug resistance in NSCLC through regulating β-catenin. Also, other HSPs, such as HSP27/HSPB1, HSP90/HSPC, HSP70/HSPA1, and GRP78/HSPA5 were also identified to be implicated in DDP resistance (35).
In exploration of the mechanism, we found that HNF1A could bind to the promoter region of DNAJC12 and served as a transcriptional activator to promote DNAJC12. Moreover, we found that HNF1A was upregulated in NSCLC tissues and showed a positive correlation with DNAJC12 expression. While HNF1A promoted aerobic glycolysis and DDP resistance in NSCLC cells, these were rescued by DNAJC12 downregulation, indicating that HNF1A promoted glycolysis and drug resistance in NSCLC cells through upregulating DNAJC12 expression. Evidence has demonstrated that HNF1A is also involved in cancer progression and drug resistance. Overexpression of HNF1A has been shown to increase the expression of stem cell markers and tumor sphere formation in pancreatic cancer cells and accelerated cell growth in primary pancreatic ductal adenocarcinoma cells (25). By inhibiting MMP14 and HNF1A expression in cervical cancer, miR-484 serves as a tumor suppressor (36). These results demonstrated that HNF1A serves as an oncogene. However, Gong et al. (37) reported that HNF1A overexpression inhibited cell growth and migration, and induced apoptosis in pancreatic cancer CanPan-1 cells. Overexpression of HNF1A enhances gemcitabine sensitivity in pancreatic ductal adenocarcinoma both in vitro and in vitro (38). The different roles of HNF1A in carcinogenesis may be caused by different cell contents.
Several limitations of this study should be clarified: the small human sample size, and we did not explore whether DNAJC12 could accelerate NSCLC progression through other mechanisms. In addition, Fang et al. (39) showed that β-catenin overexpression was closely associated with the resistance of NSCLC cells to gefitinib, an epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) used for the targeted therapy of NSCLC, indicating that DNAJC12 may also cause the resistance of NSCLC to targeted therapy. We intend to continue to explore these aspects in our next work.
In conclusion, this study revealed that DNAJC12, activated by the transcription factor HNF1A, could enhance aerobic glycolysis and drug resistance to DDP in NSCLC. It is possible that DNAJC12 is a promising treatment target for overcoming drug resistance in NSCLC. DNAJC12 is a potential candidate target for the treatment of NSCLC, and overcoming the DDP resistance.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1475/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1475/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1475/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of Shanghai General Hospital (No. 2018KY086) and informed consent was taken from all the patients. Animal experiments were performed under a project license (No. 2018AWS0095) granted by the Ethics Committee of Shanghai General Hospital, in compliance with the institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Xie Y, Zhang Y, Du L, et al. Circulating long noncoding RNA act as potential novel biomarkers for diagnosis and prognosis of non-small cell lung cancer. Mol Oncol 2018;12:648-58. [Crossref] [PubMed]
- Ardizzoni A, Boni L, Tiseo M, et al. Cisplatin- versus carboplatin-based chemotherapy in first-line treatment of advanced non-small-cell lung cancer: an individual patient data meta-analysis. J Natl Cancer Inst 2007;99:847-57. [Crossref] [PubMed]
- Song TH, Chen XX, Lee CK, et al. Dendrobine targeting JNK stress signaling to sensitize chemotoxicity of cisplatin against non-small cell lung cancer cells in vitro and in vivo. Phytomedicine 2019;53:18-27. [Crossref] [PubMed]
- Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer 2004;4:891-9. [Crossref] [PubMed]
- Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol 2011;27:441-64. [Crossref] [PubMed]
- Smolle E, Leko P, Stacher-Priehse E, et al. Distribution and prognostic significance of gluconeogenesis and glycolysis in lung cancer. Mol Oncol 2020;14:2853-67. [Crossref] [PubMed]
- Monaco L, Gemelli M, Gotuzzo I, et al. Metabolic Parameters as Biomarkers of Response to Immunotherapy and Prognosis in Non-Small Cell Lung Cancer (NSCLC): A Real World Experience. Cancers (Basel) 2021;13:1634. [Crossref] [PubMed]
- Yao J, Li R, Liu X, et al. Prognostic implication of glycolysis related gene signature in non-small cell lung cancer. J Cancer 2021;12:885-98. [Crossref] [PubMed]
- Schwartz L, Supuran CT, Alfarouk KO. The Warburg Effect and the Hallmarks of Cancer. Anticancer Agents Med Chem 2017;17:164-70. [Crossref] [PubMed]
- Vaupel P, Schmidberger H, Mayer A. The Warburg effect: essential part of metabolic reprogramming and central contributor to cancer progression. Int J Radiat Biol 2019;95:912-9. [Crossref] [PubMed]
- Marcucci F, Rumio C. Glycolysis-induced drug resistance in tumors-A response to danger signals? Neoplasia 2021;23:234-45. [Crossref] [PubMed]
- Wang X, Zhang H, Yang H, et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol 2020;14:539-55. [Crossref] [PubMed]
- Shevtsov M, Multhoff G. Heat Shock Protein-Peptide and HSP-Based Immunotherapies for the Treatment of Cancer. Front Immunol 2016;7:171. [Crossref] [PubMed]
- Straniero L, Guella I, Cilia R, et al. DNAJC12 and dopa-responsive nonprogressive parkinsonism. Ann Neurol 2017;82:640-6. [Crossref] [PubMed]
- Uno Y, Kanda M, Miwa T, et al. Increased Expression of DNAJC12 is Associated with Aggressive Phenotype of Gastric Cancer. Ann Surg Oncol 2019;26:836-44. [Crossref] [PubMed]
- He HL, Lee YE, Chen HP, et al. Overexpression of DNAJC12 predicts poor response to neoadjuvant concurrent chemoradiotherapy in patients with rectal cancer. Exp Mol Pathol 2015;98:338-45. [Crossref] [PubMed]
- Li Y, Li M, Jin F, et al. DNAJC12 promotes lung cancer growth by regulating the activation of β-catenin. Int J Mol Med 2021;47:105. [Crossref] [PubMed]
- Huang J, Tian F, Song Y, et al. A feedback circuit comprising EHD1 and 14-3-3ζ sustains β-catenin/c-Myc-mediated aerobic glycolysis and proliferation in non-small cell lung cancer. Cancer Lett 2021;520:12-25. [Crossref] [PubMed]
- Chen H, Zhang M, Zhang W, et al. Downregulation of BarH-like homeobox 2 promotes cell proliferation, migration and aerobic glycolysis through Wnt/β-catenin signaling, and predicts a poor prognosis in non-small cell lung carcinoma. Thorac Cancer 2018;9:390-9. [Crossref] [PubMed]
- Tong L, Wu W. Effects of long non-coding RNA (lncRNA) cancer susceptibility candidate 2c (CASC2c) on proliferation, metastasis and drug resistance of non-small cell lung cancer (NSCLC) cells through ERK1/2 and β-catenin signaling pathways. Pathol Res Pract 2019;215:152522. [Crossref] [PubMed]
- Tripathi SK, Biswal BK. SOX9 promotes epidermal growth factor receptor-tyrosine kinase inhibitor resistance via targeting β-catenin and epithelial to mesenchymal transition in lung cancer. Life Sci 2021;277:119608. [Crossref] [PubMed]
- Ma P, Li L, Liu F, et al. HNF1A-Induced lncRNA HCG18 Facilitates Gastric Cancer Progression by Upregulating DNAJB12 via miR-152-3p. Onco Targets Ther 2020;13:7641-52. [Crossref] [PubMed]
- Abel EV, Goto M, Magnuson B, et al. HNF1A is a novel oncogene that regulates human pancreatic cancer stem cell properties. Elife 2018;7:33947. [Crossref] [PubMed]
- Fujino S, Miyoshi N, Ito A, et al. HNF1A regulates colorectal cancer progression and drug resistance as a downstream of POU5F1. Sci Rep 2021;11:10363. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Sun B, Dong C, Lei H, et al. Propranolol inhibits proliferation and invasion of hemangioma-derived endothelial cells by suppressing the DLL4/Notch1/Akt pathway. Chem Biol Interact 2018;294:28-33. [Crossref] [PubMed]
- De Bessa SA, Salaorni S, Patrão DF, et al. JDP1 (DNAJC12/Hsp40) expression in breast cancer and its association with estrogen receptor status. Int J Mol Med 2006;17:363-7. [Crossref] [PubMed]
- Xu Q, Tu J, Dou C, et al. HSP90 promotes cell glycolysis, proliferation and inhibits apoptosis by regulating PKM2 abundance via Thr-328 phosphorylation in hepatocellular carcinoma. Mol Cancer 2017;16:178. [Crossref] [PubMed]
- Wu X, Guo J, Chen Y, et al. The 60-kDa heat shock protein regulates energy rearrangement and protein synthesis to promote proliferation of multiple myeloma cells. Br J Haematol 2020;190:741-52. [Crossref] [PubMed]
- Subramanian C, Gorney R, Wang T, et al. A novel heat shock protein inhibitor KU757 with efficacy in lenvatinib-resistant follicular thyroid cancer cells overcomes up-regulated glycolysis in drug-resistant cells in vitro. Surgery 2021;169:34-42. [Crossref] [PubMed]
- Bhattacharya B, Mohd Omar MF, Soong R. The Warburg effect and drug resistance. Br J Pharmacol 2016;173:970-9. [Crossref] [PubMed]
- Xu RH, Pelicano H, Zhou Y, et al. Inhibition of glycolysis in cancer cells: a novel strategy to overcome drug resistance associated with mitochondrial respiratory defect and hypoxia. Cancer Res 2005;65:613-21. [PubMed]
- Krawczyk Z, Gogler-Pigłowska A, Sojka DR, et al. The Role of Heat Shock Proteins in Cisplatin Resistance. Anticancer Agents Med Chem 2018;18:2093-109. [Crossref] [PubMed]
- Hu Y, Wu F, Liu Y, et al. DNMT1 recruited by EZH2-mediated silencing of miR-484 contributes to the malignancy of cervical cancer cells through MMP14 and HNF1A. Clin Epigenetics 2019;11:186. [Crossref] [PubMed]
- Gong Y, Dai HS, Shu JJ, et al. LNC00673 suppresses proliferation and metastasis of pancreatic cancer via target miR-504/ HNF1A. J Cancer 2020;11:940-8. [Crossref] [PubMed]
- Lu Y, Xu D, Peng J, et al. HNF1A inhibition induces the resistance of pancreatic cancer cells to gemcitabine by targeting ABCB1. EBioMedicine 2019;44:403-18. [Crossref] [PubMed]
- Fang X, Gu P, Zhou C, et al. β-Catenin overexpression is associated with gefitinib resistance in non-small cell lung cancer cells. Pulm Pharmacol Ther 2014;28:41-8. [Crossref] [PubMed]
(English Language Editor: J. Jones)


