
Prognostic value of CD247 in patients with head and neck squamous cell carcinoma: bioinformatic analysis of TCGA database
Introduction
With 878,348 new cases and 444,347 deaths in 2020 (1), head and neck squamous cell carcinoma (HNSC) is the 7th most common type of cancer in the world (2). Surgery, radiotherapy, chemotherapy, and immunotherapy are the main treatment methods for HNSC (3-5). More than 50% of HNSC patients develop local recurrence or metastasis in 3 years (6), and previous studies have shown that phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), tumor protein 53 (TP53), cyclin dependent kinase inhibitor 2A (CDKN2A), and other genes play an important role in the occurrence and development of HNSC (7,8). In vitro experiments in HNSC patients with PIK3CA mutations showed that PIK3CA inhibitors exhibited antitumor effects and slowed tumor cell proliferation (9). Inhibition of PIK3CA can increase the radiosensitivity of head and neck squamous cell carcinoma, hinder the DNA repair of tumor cells and improve the prognosis of HNSC patients (10). Compared with TP53 wild-type HNSC patients, tumor cells in TP53 mutant HNSC patients showed stronger invasiveness and metastasis (11). TP53 wild-type tumor cells maintain metabolic diversity, including mitochondrial respiration and glycolysis, while TP53 mutant tumor cells show glycolysis dependence, which affects the sensitivity of tumor cells to radiotherapy and thus affects patient prognosis (12). Therefore, exploring the molecular mechanism of HNSC plays a crucial role in improving the prognosis of patients. In recent years, microarray technology and bioinformatic analyses have been widely used to find gene alterations at the genome level to help identify the differentially expressed genes (DEGs) and functional pathways involved in the HNSC process. In this study, data obtained from The Cancer Genome Atlas (TCGA) underwent analyses, including a differential expression analysis and a univariate survival analysis, combined with a protein-protein interaction (PPI) analysis, Gene Ontology (GO)/Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis, clinical correlation analysis, and Gene Set Enrichment Analysis (GSEA) analysis, to explore the underlying molecular mechanisms of HNSC. We present the following article in accordance with the REMARK reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1143/rc).
Methods
The clinical and transcriptome data of the HNSC patients were acquired from TCGA (https://portal.gdc.cancer.gov/). The transcriptome data were converted to gene symbols according to Genome Reference Consortium Human Build 38 patch release 13 (GRCh38.p13, National Center for Biotechnology Information-NCBI: GCA_000001405.28). The messenger ribonucleic acid (mRNA) data were extracted, and the expression level of mRNA was measured. If there were multiple groups of mRNA data in the same specimen, the average value was used. The survival status and survival time of the patients were extracted from TCGA clinical data file, and a single-factor Cox regression analysis was performed according to the survival status and mRNA expression of each sample to generate a survival-related gene set. According to the transcriptome data, the samples were divided into a normal tissue group and a tumor tissue group, and the gene sets with differential expression in the normal tissues and tumor tissues were identified.
The intersection gene set was obtained from the survival-related gene set and the DEGs set, and the intersection gene set was imported into STRING (Protein-Protein Interaction Networks Functional Enrichment Analysis, version 11.5) (13) for the PPI analysis, and the analysis results were then imported into Cytoscape (version 3.9.0) (14) for the subsequent analysis. We used Cytoscape’s following plug-in applications: Mcode (version 1.5.1) (15) for the module analysis, Cytohubba (version 2.5.8) (16) for gene scoring, and ClueGO (17) for the gene function enrichment and pathway analysis. Based on the above analyses, CD247 molecule (CD247) was ultimately selected as the main target gene in this study.
The single-gene analysis of CD247 included a violin plot analysis, survival analysis, clinical correlation analysis, and GSEA pathway correlation analysis.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was retrospective in nature. TCGA clinical data and transcriptome data were extracted for the bioinformatic analysis. The endpoints of the study were the 5-year overall survival rate and the median survival time of the patients. The following clinical indicators were included in the analysis: age, gender, smoking history, alcohol history, tumor clinical stage, tumor pathological stage, and lymph node-related events. The transcriptome gene indicator examined in the analysis was CD247.
Statistical analysis
SPSS (version 26.0) was used for the statistical analysis, P<0.05 was considered a significant difference or a positive result. The study samples were divided into normal tissue and tumor tissue, and the expression of CD247 between the two groups was compared by independent sample t-test. The CD247 was divided into high expression group and low expression group, and Kaplan-Meier survival analysis was performed with the survival time of patients, The median survival time and 5-year overall survival rate were compared between the two groups. CD247 was divided into high expression group and low expression group, and Pearson correlation test was performed with clinical features. The clinical features correlated with CD247 expression were included in binary logistic regression analysis model for analysis. Univariate Cox regression analysis was performed on the extracted clinical feature information and patient survival time. The positive results of univariate Cox regression model were imported into multivariate Cox regression model to screen out independent prognostic factors of HNSC patients.
Results
Volcano plot and Venn diagram
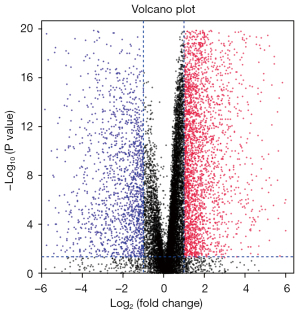
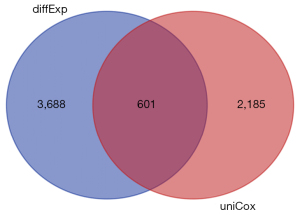
We downloaded the TCGA transcriptome expression data, and through a differential expression analysis, the cutoff value was defined as a log fold change (FC) with an absolute value >1, and a P value <0.05. The distribution trend of the included genes is shown in Figure 1, and a total of 4,289 DEGs were counted. TCGA clinical data and transcriptome data were extracted, a univariate Cox regression analysis was performed, and a total of 2,786 survival-related genes were identified. Using the Venn diagram (https://bioinformatics.psb.ugent.be/webtools/Venn/) to identify the intersection (see Figure 2), a total of 601 intersection genes were identified as the object of the subsequent analysis.
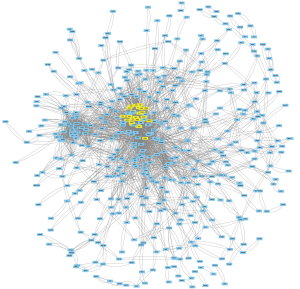
Cytoscape overview map and Multi-Contrast Delayed Enhancement (MCODE) key modules
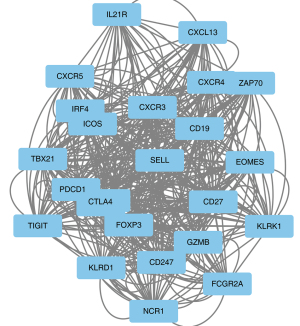
The identified intersection genes were imported into STRING for the PPI analysis, the parameter was set to medium confidence (0.400), and the analysis results were imported into Cytoscape for visualization. A module analysis was conducted via MCODE. Figure 3 shows an overview of Cytoscape. Figure 4 shows the module with the highest MCODE score (score =19.304) with a total of 24 hub genes.
A functional enrichment analysis of the intersection genes was carried out, and the similar results of functional enrichment analysis were take only one result. The analysis results were visualized in a bubble diagram (see Figure 5). Notably, the main cellular components of the intersection genes were the endoplasmic reticulum lumen and the intrinsic component of the plasma membrane. The molecular functions were signaling receptor binding and oxidoreductase activity. The main biological processes included the cell surface receptor signaling pathway, leukocyte activation, cell motility, lymphocyte activation, and lymph node development.
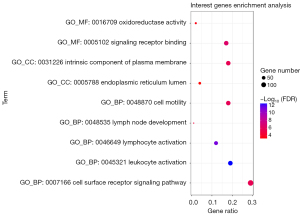
Pathways involved in the hub genes and CD247 protein interaction score
The results of the functional enrichment analysis of the hub genes by ClueGO are shown in Table 1. Notably, the hub genes act on multiple immune-related pathways, including the T-helper 17 (Th17) cell differentiation, natural killer cell mediated cytotoxicity, the T cell receptor signaling, the viral protein interaction with the cytokine and cytokine receptor, and the primary immunodeficiency pathways. Among them, CD247 was found to be involved in multiple pathways, which suggests that it may play an important role in immune regulation. To further assess the importance of CD247, the PPIs were scored using Cytohubba. Using the Maximal Clique Centrality (MCC) scoring method, the results showed that the CD247 interaction score was in the top 3%, which suggests that it plays an important role at the protein level. Thus, CD247 was selected as the main target gene in this study.
Table 1
| Term | FDR | Genes | Associated genes found |
|---|---|---|---|
| KEGG:04659 Th17 cell differentiation | 3.12E-06 | 6/24 | [CD247, FOXP3, IL21R, IRF4, TBX21, ZAP70] |
| KEGG:04650 Natural killer cell mediated cytotoxicity | 9.04E-06 | 6/24 | [CD247, GZMB, KLRD1, KLRK1, NCR1, ZAP70] |
| KEGG:04660 T cell receptor signaling pathway | 5.34E-05 | 5/24 | [CD247, CTLA4, ICOS, PDCD1, ZAP70] |
| KEGG:04061 Viral protein interaction with cytokine and cytokine receptor | 6.49E-04 | 4/24 | [CXCL13, CXCR3, CXCR4, CXCR5] |
| KEGG:05340 Primary immunodeficiency | 7.93E-04 | 3/24 | [CD19, ICOS, ZAP70] |
KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially expressed genes; FDR, false discovery rate.
Single-gene CD247 analysis
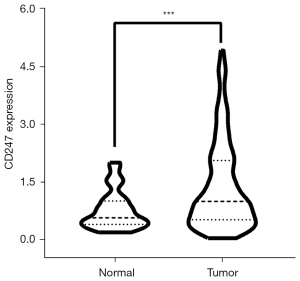
TCGA transcriptome data were extracted to draw a violin plot (see Figure 6). The violin plot suggested that CD247 was more highly expressed in tumor tissues than normal tissues, and there was a significant difference between the 2 groups. The median CD247 expression value for the normal group was 0.559, and that for the tumor group was 0.985. The independent samples t-test showed a P value <0.001, and a t value of 6.647.
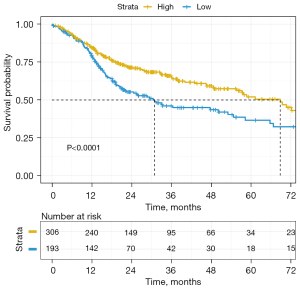
TCGA clinical data were used to draw a survival curve graph (see Figure 7). The optimal cutoff value of 0.816 was determined by R survminer (Kassambara, Kosinski, & Biecek, 2021). The survival curves were drawn using ‘ggplot2’. R package (version 0.4.9), and the CD247 data were divided into a high-expression group and a low-expression group. Kaplan-Meier survival curves were drawn, and there was a significant difference between the 2 groups (P<0.0001, χ2=13.061). The median survival time of the low-expression group was 30.9 months, and the 5-year survival rate was 36.4%. While the median survival time of the high-expression group was 68.8 months, and the 5-year survival rate was 52.3%. Thus, patients with high CD247 expression appeared to have a better prognosis than patients with low CD247 expression.
As Table 2 shows, based on the best CD247 cutoff, the patients were divided into a high-expression group and a low-expression group, and the correlation analysis of TCGA clinical data showed that CD247 expression was positively correlated with patient age (P=0.005), negatively correlated with pathological tumor stage (pT) (P=0.011), and negatively correlated with lymph node extracapsular invasion (P=0.014). We incorporated the above-mentioned related factors into a binary logistic regression analysis and found that the risk of developing the disease increased by 3.5% for each 1-year increase in age. Additionally, the risk of developing pT4 stage in the high-expression group of CD247 was 36.5% of that in the low-expression group. Thus, the high expression of CD247 appears to play a role in reducing the pathological tumor stage.
Table 2
| Parameter | n | r | P | Exp(B) | 95% CI | P |
|---|---|---|---|---|---|---|
| Age | 499 | 0.127 | 0.005 | 1.035 | 1.010–1.062 | 0.007 |
| Gender | 500 | 0.040 | 0.368 | |||
| Alcohol history | 489 | −0.043 | 0.348 | |||
| Tobacco smoking history | 490 | 0.027 | 0.544 | |||
| Clinical | ||||||
| Stage (I vs. IV) | 294 | –0.059 | 0.316 | |||
| cT (1 vs. 4) | 215 | –0.112 | 0.102 | |||
| cN | 481 | 0.071 | 0.120 | |||
| M | 480 | 0.038 | 0.403 | |||
| Pathological | ||||||
| Grade (I vs. IV) | 182 | 0.128 | 0.085 | |||
| pT (1 vs. 4) | 213 | –0.174 | 0.011 | 0.365 | 0.169–0.789 | 0.010 |
| pN | 403 | –0.047 | 0.349 | |||
| Lymphovascular invasion present | 339 | –0.058 | 0.290 | |||
| Perineural invasion present | 351 | –0.090 | 0.093 | |||
| Pathological nodal extracapsular spread | 347 | –0.131 | 0.014 |
Categorical dependent variable, greater or less than the cutoff level. r, Pearson correlation coefficient; CI, confidence interval.
The univariate Cox regression analysis showed that age (P<0.001), clinical lymph node stage (N stage) (P=0.035), clinical distant metastasis stage (M stage) (P=0.003), new tumor events (P=0.001), and CD247 (P=0.014) were associated with overall survival in HNSC patients (see Table 3). The results of the univariate Cox regression analysis were included in the multivariate Cox regression analysis, and we found that each 1-year increase in age was associated with a 3% increase in the risk of onset [P<0.001, confidence interval (CI): 1.01–1.04]. Further, a 1-level increase in the clinical lymph node stage (N stage) resulted in a 29% increase in the risk of onset (P=0.001, CI: 1.10–1.51). Additionally, with each level increase in the clinical metastasis stage (M stage), the risk of onset increased by 498% (P=0.001, CI: 2.17–16.45), and the risk of new tumor events increased by 110% (P<0.001, CI: 1.59–2.78). In the high CD247 expression group, the risk of onset was reduced by 26% (P=0.002, CI: 0.60–0.90).
Table 3
| Parameter | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | ||
| Age | 1.02 | 1.01–1.04 | 0.000 | 1.03 | 1.01–1.04 | 0.000 | |
| Gender | 1.28 | 0.95–1.72 | 0.106 | ||||
| Alcohol history | 1.00 | 0.74–1.34 | 0.984 | ||||
| Smoking history | 0.98 | 0.87–1.11 | 0.740 | ||||
| Clinical stage | 1.12 | 0.95–1.31 | 0.173 | ||||
| cT | 1.10 | 0.95–1.27 | 0.221 | ||||
| cN | 1.18 | 1.01–1.37 | 0.035 | 1.29 | 1.10–1.51 | 0.001 | |
| M | 4.56 | 1.68–12.39 | 0.003 | 5.98 | 2.17–16.45 | 0.001 | |
| Grade | 1.11 | 0.90–1.38 | 0.330 | ||||
| New tumor events | 1.31 | 1.12–1.53 | 0.001 | 2.10 | 1.59–2.78 | 0.000 | |
| CD247 | 0.78 | 0.64–0.95 | 0.014 | 0.74 | 0.60–0.90 | 0.002 | |
OS, overall survival; HNSC, head and neck squamous cell carcinoma; HR, hazard ratio; CI, confidence interval.
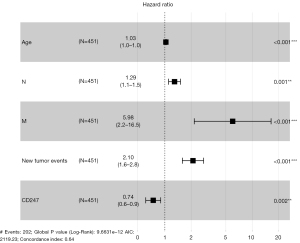
The results of the multivariate Cox regression analysis were plotted as a forest plot (see Figure 8), which showed that age, CD247, N, M, and new tumor events were independent risk factors in the prognosis of HNSC patients. Among them, CD247 was found to be a protective factor, and patients with high CD247 expression had a better prognosis than those with low CD247 expression.
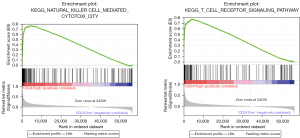
TCGA transcriptome data were imported into GSEA for a pathway analysis (see Figure 9). The GSEA analysis showed that the expression of CD247 was significantly correlated with the activation of hsa04650 natural killer cell mediated cytotoxicity and the hsa04660 T cell receptor signaling pathway (hsa04650: NSE =2.613513, false discovery rate (FDR)-q =0.001<0.25; hsa04660: NSE =2.610915, FDR-q =0.001<0.25). Thus, the high expression of CD247 appears to activate these 2 pathways.
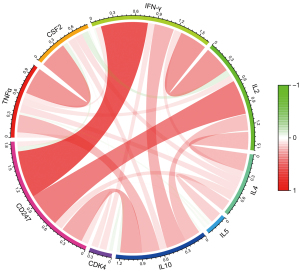
Further examinations of the pathways revealed that hsa04650 was involved in changes in the levels of tumor necrosis factor alpha (TNFα), colony stimulating factor 2 (CSF2), and interferon gamma (IFN-γ) in the nucleus, while hsa04660 was involved in expression changes in the levels of interleukin (IL)-2, IL-4, IL5, IL10, TNFα, CSF2, IFN-γ, and cyclin dependent kinase 4 (CDK4). The above-mentioned related pathway indicators were extracted from TCGA transcriptome data, and their correlation with CD247 was analyzed and plotted (see Figure 10). In hsa04650 CD247 was found to be highly positively correlated with IFN-γ (r=0.664, P<0.001). Additionally, in hsa04660, CD247 was found to be highly positively correlated with IFN-γ (r=0.664, P<0.001), and moderately positively correlated with IL-2 (r=0.487, P<0.001), and IL-10 (r=0.309, P<0.001).
Discussion
In previous studies, we found that IFN-γ, which is a type II interferon produced by immune cells, such as T cells and natural killer cells (18,19). In our study, it is a related indicator of the 2 pathways. IFN-γ activates effector immune cells and enhances antigen presentation. It plays an important role in antiviral and anti-tumor responses (18,19), and participates in the class I antigen presentation pathway by inducing the replacement of the catalytic proteasome subunit with the immunoproteasome subunit (18,19). IFN sensitization can improve the anti-tumor effect of chimeric antigen receptor T-cell immunotherapy (CAR-T) (20). Changes in leukocyte morphology and behavior caused by IL-2 lead to the initiation or continuation of immune responses (21), and mediate immune responses (22). IL-2 is involved in intercellular signal transduction and plays a role in activating natural killer cells (23). IL-2 can also transmit signals from the cell surface to trigger apoptosis (24,25), and is involved in processes that activate or increase the frequency, rate, or extent of interferon-gamma production (26). IL-10 plays a role in regulating leukocyte chemotaxis (27), and the transduction of IL-10 promotes the maintenance of the programmed cell death protein 1 (PD-1), T cell factor-1 (TCF-1), cytotoxic T lymphocyte (CD8 T) cell population, thereby maintaining anti-tumor immunity (28,29). In-vivo experiments have shown that the overexpression of IL-10 improves tumor immune monitoring ability and inhibits tumor growth (30). In clinical practice, polyethylene glycol IL-10 has been shown to result in immune activation and has anti-tumor effects in patients with advanced tumors (31).
In summary, IFN-γ, IL-2, and IL-10 may produce anti-tumor effects through various ways. In our study, CD247 had a moderate-to-high positive correlation with the expression of IFN-γ, IL-2, and IL-10. In the multivariate Cox regression analysis, the high expression of CD247 was shown to be a protective factor in the prognosis of HNSC patients.
In the clinical correlation analysis, the patients were divided into high- and low-expression groups based on the best cutoff value. The correlation analysis of TCGA clinical data revealed a positive correlation between CD247 expression and patient age (P=0.005). This phenomenon may be associated to multiple chronic inflammation in elderly patients (32,33). CD247 was negatively correlated with pT stage (P=0.011) and pathological nodal extracapsular spread (P=0.014). Based on these clinical indicators and the subsequent pathway analysis results, we speculate that CD247 promotes the expression of IFN-γ, IL-2, and IL-10 by activating hsa04650 and hsa04660, thereby improving the body’s tumor immune monitoring ability, inducing tumor cell apoptosis, and inhibiting tumor cell growth. This process clinically manifests as a reduction in the pT stage and a reduction in the occurrence of pathological nodal extracapsular spread, which in turn improve the prognosis of HNSC patients.
CD247 can be considered a protective factor in the prognosis of HNSC patients. Increasing the expression of CD247 may improve the prognosis of HNSC patients. It is a potential target for improving the clinical treatment and prognosis of HNSC patients. However, further research needs to be conducted to confirm this conclusion.
Conclusions
Through our research, it can be inferred that in HNSC patients, the high expression of CD247 increases the expression levels of IFN-γ, IL-2, and IL-10 in the nucleus by activating the hsa04650 and hsa04660 pathways, which in turn improves tumor immune monitoring, promotes tumor cell apoptosis and inhibits tumor cell growth. This process may reduce the pT stage of patients and reduce the occurrence of pathological nodal extracapsular spread events, thereby improving the prognosis of patients. Thus, CD247 may serve as a potential target for improving clinical treatments or improving prognosis.
Acknowledgments
Funding: The study was supported by Guangzhou Health Science and Technology Project (grant No. 20201A011101).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1143/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1143/coif). The authors report that this study was supported by Guangzhou Health Science and Technology Project (grant No. 20201A011101). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Mody MD, Rocco JW, Yom SS, et al. Head and neck cancer. Lancet 2021;398:2289-99. [Crossref] [PubMed]
- Johnson DE, Burtness B, Leemans CR, et al. Head and neck squamous cell carcinoma. Nat Rev Dis Primers 2020;6:92. [Crossref] [PubMed]
- Carlisle JW, Steuer CE, Owonikoko TK, et al. An update on the immune landscape in lung and head and neck cancers. CA Cancer J Clin 2020;70:505-17. [Crossref] [PubMed]
- Karam SD, Raben D. Radioimmunotherapy for the treatment of head and neck cancer. Lancet Oncol 2019;20:e404-16. [Crossref] [PubMed]
- Sacco AG, Cohen EE. Current Treatment Options for Recurrent or Metastatic Head and Neck Squamous Cell Carcinoma. J Clin Oncol 2015;33:3305-13. [Crossref] [PubMed]
- Guan Y, Wang G, Fails D, et al. Unraveling cancer lineage drivers in squamous cell carcinomas. Pharmacol Ther 2020;206:107448. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576-82. [Crossref] [PubMed]
- Jin N, Keam B, Cho J, et al. Therapeutic implications of activating noncanonical PIK3CA mutations in head and neck squamous cell carcinoma. J Clin Invest 2021;131:e150335. [Crossref] [PubMed]
- Zumsteg ZS, Morse N, Krigsfeld G, et al. Taselisib (GDC-0032), a Potent β-Sparing Small Molecule Inhibitor of PI3K, Radiosensitizes Head and Neck Squamous Carcinomas Containing Activating PIK3CA Alterations. Clin Cancer Res 2016;22:2009-19. [Crossref] [PubMed]
- Sano D, Xie T, Ow T, et al. Disruptive TP53 mutation is associated with aggressive disease characteristics in an orthotopic murine model of oral tongue cancer. Clin Cancer Res 2011;17:6658-70. [Crossref] [PubMed]
- Wilkie M, Lau A, Vlatkovic N, et al. Tumour metabolism in squamous cell carcinoma of the head and neck: an in-vitro study of the consequences of TP53 mutation and therapeutic implications. Lancet 2015;385:S101. [Crossref] [PubMed]
- Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 2013;41:D808-15. [Crossref] [PubMed]
- Smoot ME, Ono K, Ruscheinski J, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 2011;27:431-2. [Crossref] [PubMed]
- Bandettini WP, Kellman P, Mancini C, et al. MultiContrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: a clinical validation study. J Cardiovasc Magn Reson 2012;14:83. [Crossref] [PubMed]
- Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8:S11. [Crossref] [PubMed]
- Bindea G, Mlecnik B, Hackl H, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009;25:1091-3. [Crossref] [PubMed]
- Hisamatsu H, Shimbara N, Saito Y, et al. Newly identified pair of proteasomal subunits regulated reciprocally by interferon gamma. J Exp Med 1996;183:1807-16. [Crossref] [PubMed]
- El Bougrini J, Pampin M, Chelbi-Alix MK. Arsenic enhances the apoptosis induced by interferon gamma: key role of IRF-1. Cell Mol Biol (Noisy-le-grand) 2006;52:9-15. [PubMed]
- Dong E, Yue XZ, Shui L, et al. IFN-γ surmounts PD-L1/PD1 inhibition to CAR-T cell therapy by upregulating ICAM-1 on tumor cells. Signal Transduct Target Ther 2021;6:20. [Crossref] [PubMed]
- Feng Y, Huang N, Wu Q, et al. HMGN2: a novel antimicrobial effector molecule of human mononuclear leukocytes? J Leukoc Biol 2005;78:1136-41. [Crossref] [PubMed]
- Weinberg K, Parkman R. Severe combined immunodeficiency due to a specific defect in the production of interleukin-2. N Engl J Med 1990;322:1718-23. [Crossref] [PubMed]
- Minami Y, Kono T, Miyazaki T, et al. The IL-2 receptor complex: its structure, function, and target genes. Annu Rev Immunol 1993;11:245-68. [Crossref] [PubMed]
- Bredesen DE, Mehlen P, Rabizadeh S. Apoptosis and dependence receptors: a molecular basis for cellular addiction. Physiol Rev 2004;84:411-30. [Crossref] [PubMed]
- Mason EF, Rathmell JC. Cell metabolism: an essential link between cell growth and apoptosis. Biochim Biophys Acta 2011;1813:645-54. [Crossref] [PubMed]
- Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev 2004;202:8-32. [Crossref] [PubMed]
- Gesser B, Leffers H, Jinquan T, et al. Identification of functional domains on human interleukin 10. Proc Natl Acad Sci U S A 1997;94:14620-5. [Crossref] [PubMed]
- Guo Y, Xie YQ, Gao M, et al. Metabolic reprogramming of terminally exhausted CD8+ T cells by IL-10 enhances anti-tumor immunity. Nat Immunol 2021;22:746-56. [Crossref] [PubMed]
- Hanna BS, Llaó-Cid L, Iskar M, et al. Interleukin-10 receptor signaling promotes the maintenance of a PD-1int TCF-1+ CD8+ T cell population that sustains anti-tumor immunity. Immunity 2021;54:2825-41.e10. [Crossref] [PubMed]
- Mumm JB, Emmerich J, Zhang X, et al. IL-10 elicits IFNγ-dependent tumor immune surveillance. Cancer Cell 2011;20:781-96. [Crossref] [PubMed]
- Naing A, Papadopoulos KP, Autio KA, et al. Safety, Antitumor Activity, and Immune Activation of Pegylated Recombinant Human Interleukin-10 (AM0010) in Patients With Advanced Solid Tumors. J Clin Oncol 2016;34:3562-9. [Crossref] [PubMed]
- Bulati M, Caruso C, Colonna-Romano G. From lymphopoiesis to plasma cells differentiation, the age-related modifications of B cell compartment are influenced by "inflamm-ageing". Ageing Res Rev 2017;36:125-36. [Crossref] [PubMed]
- Cevenini E, Monti D, Franceschi C. Inflamm-ageing. Curr Opin Clin Nutr Metab Care 2013;16:14-20. [Crossref] [PubMed]


