Heterogeneous microenvironment analysis to explore the potential regulatory role of endothelial-mesenchymal transition in idiopathic pulmonary fibrosis
Introduction
Idiopathic pulmonary fibrosis (IPF) is a chronic, irreversible, and fatal lung disease, which is characterized by fibroblast proliferation and excessive deposition of extracellular matrix (ECM) in lung tissue (1). With the development of the disease, lung function gradually declines, leading to respiratory failure and even death. The median survival period after initial diagnosis is 2–3 years (2,3). Clinically, lung transplantation is an effective therapeutic method to improve the quality of life and survival rate of patients with IPF (4). However, given the short supply of donor organs and some serious immune rejection responses after transplantations, only a small fraction of the patients can benefit from lung transplantation. In addition, nintedanib and pirfenidone have been approved to treat IPF by the US Food and Drug Administration (5). However, the therapeutic effect of these drugs is limited, and is accompanied by multiple adverse reactions (6,7). Therefore, there is a pressing need to better understand the underlying pathogenesis and molecular mechanism of IPF, in order to explore new treatment means (8).
The tumor microenvironment (TME) is a novel foundation inside in the bodies of tumor patients. The structure of a typical TME is composed of fibroblasts, myofibroblasts, endothelial cells, pericytes, adipocytes, immune and inflammatory cells, and ECM, which can directly affect the growth, migration, and differentiation of cancer cells (9,10). Normally, the stroma maintains a stable environment within tissue and acts as a barrier to tumor formation. However, when cells become cancerous, the surrounding matrix changes to support cancer development. Likewise, the development of fibrotic disease is a result of the interaction of multiple cell subsets with distinct genetic and phenotypic characteristics (11). Changes in cellular diversity contribute to pathological pulmonary fibrosis (12). In the TME, among fibroblasts, pericytes, and bone marrow-derived mesenchymal cells, endothelial cells adopt cancer-associated fibroblast (CAF) phenotypes through the endothelial-mesenchymal transition (EndMT) process (13). It has been reported that mesenchymal cells, immune cells, and endothelial cells synergistically regulate the production, degradation, deposition, and remodeling of the ECM, leading to the development of liver fibrosis. Interactions between fibrotic components create a unique ‘fibrotic niche’ microenvironment (14). A previous study has shown that M1 bone marrow-derived macrophages (BMDMs) recruit and modify the activation of endogenous macrophages and natural killer (NK) cells by regulating the liver microenvironment, possibly leading to hepatic stellate cell (HSC) apoptosis, thereby hindering liver fibrosis (15). Furthermore, it has also been reported that the transformation of bone marrow-derived monocytes to fibroblasts is a key step in the pathogenesis of renal fibrosis and is regulated by the inflammatory microenvironment. The axis of cluster of differentiation eight positive (CD8+) T cell and interferon (IFN)-γ-CD4+ T cell as an important microenvironment for the monocyte-to-fibroblast transition, which negatively regulates renal fibrosis (16). However, the direct link between microenvironments and IPF has not yet been confirmed. Thus, there is a pressing need to explore the connections between different cell types in the IPF microenvironment to better understand the underlying pathogenesis and molecular mechanism of IPF.
Transcriptomics is increasingly used to explore disease mechanisms. Through transcriptomics and next-generation sequencing, we can obtain an unprecedented level of detail in understanding cell phenotype and function by investigating genes expressed in specific physiological and pathological states (17). The increased availability of well characterized human tissues and the emergence of high-throughput transcriptome analysis technology have promoted a new era of IPF research. Transcriptome research has revealed many new molecules and pathways highly related to the pathogenesis of IPF, including the role of matrix metalloproteinase (MMP), developmental pathway, micro ribonucleic acid (RNA) and the importance of alveolar epithelial and myofibroblast regulatory networks in IPF. At the same time, using single cell and tissue microenvironment to improve the cellular and spatial resolution of transcriptomics is very important to study the occurrence of IPF (18). In this study, we explored the composition of the IPF microenvironment and the potential interactions of its subcellular populations by analyzing IPF single-cell sequencing data and transcriptome gene expression data.
In the past, there have been many studies on IPF, but the research on pulmonary microenvironment is still rare. However, the existing bioinformatics articles simply explore the pathogenesis of IPF through the transcriptome characteristics of large tissue homogenate, failing to capture the complexity of microenvironment. Our research starts from the heterogeneous microenvironment of IPF, and uses a unique perspective to understand how cells interact in the remodeled heterogeneous microenvironment of IPF through the changes of cell number and phenotype. We aim to discuss the pathogenic mechanism of IPF from a novel perspective, and to lay a theoretical foundation and provide data support for drug development and the discovery of pathological triggers. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1438/rc).
Methods
IPF related data set acquisition and pre-processing
The Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) is an international public database that provides high-throughput microarray and next-generation sequence-functional genomic datasets submitted by research communities (19). We obtained three IPF-related gene expression profile datasets from the GEO database: GSE150910, GSE92592, and GSE110147 (20-22), and the corresponding clinical information of each dataset is listed in Table 1. We downloaded the original data files and experimental information files of the above datasets. The GSE150910 dataset contains 103 IPF samples, 103 control samples, and the GSE92592 dataset contains 20 IPF samples and 19 control samples. The reference genomes for transcriptome sequencing of the GSE150910 and GSE92592 datasets are human gencode.v27 and human hg19, respectively. We used R4.0.3 software (http://cran.r-project.org) combined with the GenomicFeatures package (23) to annotate the gene names of the two different versions. The GSE110147 dataset contains 22 IPF samples and 11 control samples. We used R4.0.3 software to annotate the probe expression profile into the expression profile corresponding to the gene name, and the median expression of multiple probes corresponding to a single gene name was taken.
Table 1
| Characteristics | GSE150910 | GSE110147 | GSE92592 |
|---|---|---|---|
| Age (years) | |||
| <60 | 88 | NA | NA |
| ≥60 | 117 | NA | NA |
| NA | 1 | NA | NA |
| Gender | |||
| Male | 102 | NA | NA |
| Female | 104 | NA | NA |
| Sample size | |||
| Total | 206 | 33 | 39 |
| IPF | 103 | 22 | 20 |
| Control | 103 | 11 | 19 |
| Smoke | |||
| Yes | 105 | NA | NA |
| No | 83 | NA | NA |
| NA | 18 | NA | NA |
IPF, idiopathic pulmonary fibrosis; NA, not available.
Single-cell RNA sequencing data acquisition and pre-processing
We obtained the single-cell RNA sequencing dataset, GSE135893 (12), from the GEO database, and used the Seurat package in the R4.0.3 software to read the single-cell sequencing sample data for quality control (quality control threshold: nFeature_RNA >1,000, and mitochondrial-related genes account for less than 25%). The total expression level of each cell in the corrected data was further normalized to 10,000, and logarithmic normalization was subsequently performed. The normalized data were subjected to the identification of genes with highly variable expression between cells to obtain 2,000 genes. We used the ScaleData function to perform linear regression scaling based on the 2,000 highly variable genes, and then applied the principal component analysis (PCA) method for dimensionality reduction analysis. We further combined the harmony package to correct for batch effects between different samples in the PCA dimensionality reduction analysis results. The FindNeighbors function was used to construct a nearest-neighbor graph of the dimensionality reduction analysis results after eliminating the batch effect, which was then clustered, and the results were visualized.
IPF microenvironment analysis
Bulk gene expression
The xCell package (24) in R4.0.3 software was used to calculate the microenvironmental cell fractions of both the IPF and control samples for the three gene expression profiles, respectively. The combined the ggpubr package was applied to visualize the fractions with boxplots. P<0.05 was considered to indicate statistically significant differences.
Single-cell gene expression
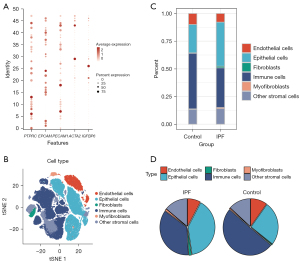
The Seurat package (25) in R4.0.3 software was used to combine protein tyrosine phosphatase receptor type C (PTPRC), epithelial cell adhesion molecule (EPCAM), platelet and endothelial cell adhesion molecule 1 (PECAM1), actin alpha 2, smooth muscle (ACTA2), insulin-like growth factor binding protein 6 (IGFBP6), and other cell markers, and annotate the above single-cell clustering results as immune cells, epithelial cells, endothelial cells, myofibroblasts, fibroblasts, and other stromal cells, respectively. All of the above proteins were confirmed to be specific biomarkers of each cell type (26-30), and the proportions of these cells in the IPF and control groups were visualized using the ggplot2 package (31).
IPF differentially expressed genes (DEGs) and enrichment analysis
To identify DEGs between the IPF and control groups, we performed DEGs analysis on the GSE150910 dataset, with the largest sample size in the gene expression profile analyzed using the limma package (32) in R4.0.3 software [DEGs screening threshold: P<0.05 after correction by Benjamini-Hochberg (BH) method, |logFC| >1]. We then employed the gseGO function of the clusterProfiler package to perform gene set enrichment analysis (GSEA) (33) on the obtained DEGs combined with the BPs (Biological Processes) background in a logFC descending order (result screening threshold: P<0.05). Finally, BPs related to endothelial cells and fibroblasts in the enrichment results were visualized.
Correlation between IPF endothelial cells and multi-scale embedded gene co-expression network analysis (MEGENA)
In this study, we used R4.0.3 software combined with the cor.test function to analyze the Pearson’s correlation between the endothelial cell score and all GSE150910 dataset gene expression values. The screening threshold was as follows: P<0.05 and the absolute value of the correlation >0.3. We then used the MEGENA package (34) to perform multi-scale chimeric gene co-expression network analysis on the gene expression profiles corresponding to the screened genes, and identified the hub genes of the co-expression module as well as its sub-modules with the largest number of genes. Finally, we investigated their expression in the GSE150910 dataset.
Transcription factors (TFs) analysis
The upstream transcription factors of the hub genes were searched in ChIPBase v2.0 (https://rna.sysu.edu.cn/chipbase/), and cytoscape 3.9.0 (www.cytoscape.org/) was used to draw the transcription factor regulatory network corresponding to the hub genes.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Statistical analysis
In this study, the limma package was used for differential gene analysis, the difference significance between groups was calculated by Bayes corrected t-test, and then the difference significance P value was corrected by BH method. Correlation analysis was performed using Pearson. R4.0.3 software was used for statistical analysis, and P<0.05 was considered statistically significant.
Results
IPF microenvironment analysis
Advances in single-cell and transcriptome sequencing have facilitated exploration of biological systems with unprecedented resolution. Firstly, the composition of cell types from lung tissues of healthy control and IPF patients in the GSE150910 (n=206), GSE110147 (n=33), and GSE92592 (n=39) datasets was analyzed to identify and construct developmental ‘trajectories’ for lung fibrosis. Compared with the control group, there were a total of 20 different cell types with altered cell scores in the IPF group, including lymphoids (B-cells, CD8+ T-cells, CD8+ central memory T-cells (Tcm), class-switched memory B-cells, memory B-cells, naive B-cells, plasma cells, type2 T-helper (Th2) cells), stem cells (platelets), myeloids (activated dendritic cells (aDC), dendritic cells (DC), immature dendritic cells (iDC), macrophages M1, monocytes, neutrophils), and stromal cells (chondrocytes, endothelial cells, fibroblasts, lymphatic (ly) endothelial cells, microvascular (mv) endothelial cells), as shown in Figure 1. Given that the relationship between immune-inflammatory cell (lymphoids and myeloids) changes and pulmonary fibrosis in transcriptome sequencing has been reported previously (35,36), we explored the underlying relationship between the changes to other cell types. As shown in Figure 1A-1C, there were four types of cell scores increased in all three datasets, including fibroblasts, B-cells, chondrocytes, and plasma cells. Interestingly, the scores of endothelial cells, ly endothelial cells, and mv endothelial cells were significantly reduced. In addition, the amount of epithelial cells exhibited different change tendencies in these three datasets.
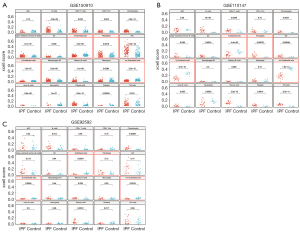
EndMT is a process in which endothelial cells lose their specific markers and obtain mesenchymal cell phenotype. This process is accompanied by the reduction of endothelial cells, leading to the proliferation and activation of fibroblasts (37). The fibroblast scores increased in all three datasets. We analyzed the cell fraction changes in the microenvironment through transcriptome sequencing data, which indicated that EndMT might occur during IPF.
Analysis of IPF microenvironment in single-cell sequencing data
In order to verify the hypothesis of IPF-associated EndMT, we further selected a single-cell sequencing dataset (GSE135893) to investigate the cellular heterogeneity and identify different cell populations. We performed annotation analysis on 33,042 cells from lung tissues of healthy control patients and 60,217 cells from lung tissues of IPF patients (total =93,259 cells). The cells were divided into five types, including immune cells, epithelial cells, endothelial cells, myofibroblasts, and fibroblasts (Figure 2A,2B), which were labeled with biomarkers including CPTPRC, EPCAM, PECAM1, ACTA2, and IGFBP6. Furthermore, we used the ggplot2 package to visualize the proportions of the aforementioned cells in the control and IPF groups, respectively. As shown in Figure 2C,2D, endothelial cells accounted for 9.9% in the control group, but decreased to 7.8% in the IPF group. Meanwhile, the fraction of fibroblasts obviously increased from 0.48% to 1.6% in the IPF group. In addition, the down-regulated proportion of endothelial cells was highly correlated to the increased amplitude of fibroblasts in IPF, and the proportion of myofibroblasts in the IPF group was also increased from 0.6% to 1.1%. By calculating the proportion of cells using single-cell sequencing data, we reported that EndMT occurred during pulmonary fibrosis.
IPF DEGs and enrichment analysis
In order to explore the correlation between gene regulatory functions and EndMT in the IPF microenvironment, we first used the dataset with the largest sample size, GSE150910 (NIPF=103, NCtrl=103), to identify the DEGs. As shown in Figure 3A,3B, the results showed a total of 327 up-regulated genes and 187 down-regulated genes in the IPF group. We further used these DEGs for GSEA enrichment analysis to reveal changes in BPs associated with endothelial cells and fibroblasts. The control group mainly exhibited correlations with endothelial cell migration and differentiation, including blood vessel endothelial cell migration, endothelial cell differentiation, and negative regulation of endothelial cell proliferation (Figure 3C). Meanwhile, BPs related to fibroblast function, including the cellular response to fibroblast growth factor stimulus and collagen fibril organization deposition, were mainly enriched in the IPF group (Figure 3D).
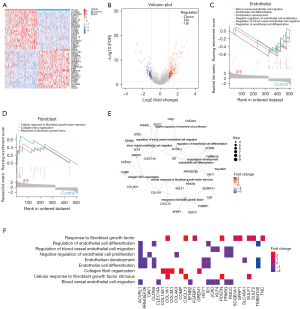
Based on the enrichment results, a functional enrichment network map and heat map were constructed to investigate genes that may regulate the BPs of endothelial cells and fibroblasts, as shown in Figure 3E,3F. We found 15 genes that were enriched in endothelial cell-related BPs, including ACVRL1, CLDN5, TMEM100, SLC40A1, ARHGEF26, CLEC14A, KLF4, EFNB2, JCAD, PPARG, RGCC, CAV1, COL4A1, HEY1, and ID1, indicating that they may play an important role in regulating endothelial cell migration and differentiation. Also, there were 13 genes enriched in fibroblast-related BPs, including COL1A1, CXCL13, POSTN, SULF1, COL14A1, COMP, COL3A1, GREM1, SFRP1, SULF2, FGFBP1, TNC, and SCGB1A1, which may be involved in the regulation of fibroblast activation and collagen deposition. The above results suggested that in the occurrence and development of IPF, genes enriched in endothelial cell BPs may promote endothelial cell function and phenotype changes by regulating endothelial cell migration and differentiation, leading to fibroblast proliferation, further transformation into myofibroblasts (mesenchymal cells), and collagen deposition.
Analysis of MEGENA co-expression network and expression of hub genes
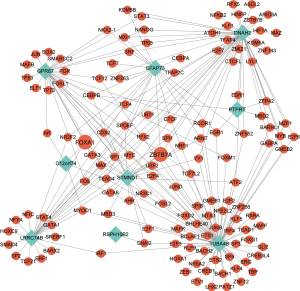
To further dissect the complex cooperative regulatory network between genes regulating endothelial cell function and phenotype in the IPF microenvironment, we used the cor.test function to analyze the Pearson correlation between endothelial cell fraction and all gene expression values in the GSE150910 dataset. A co-expression network map with multiple modules was obtained using MEGENA (Figure 4A). The network diagram consisted of 21 modules of different sizes, of which the largest was the No. 4 module, and CFAP73 and LRRC74B were the key nodes of this module. Module 4 contains six sub-modules, including c1_11, c1_12, c1_13, c1_14, c1_15, and c1_16.
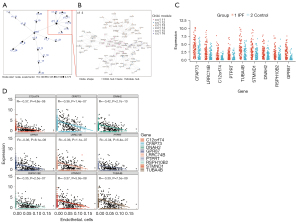
We further constructed an enlarged map of the co-expression network of module 4 (containing 98 genes) and its sub-modules (Figure 4B), and screened a total of nine hub genes, including CFAP73, LRRC74B, C12orf74, PTPRT, TUBA4B, STMND1, DNAH2, RSPH10B2, and GPR87. To explore the role of hub genes in IPF and their relationship with endothelial cells, we investigated the expression of these nine hub genes in IPF in the GSE150910 dataset, and found that the expressions of these nine hub genes was significantly up-regulated in the IPF group, which were negatively correlated with endothelial cell scores (Figure 4C,4D). From this, we inferred that hub genes may be involved in regulating the endothelial cell function and phenotype changes in the IPF microenvironment, and their expressions may play an important role in EndMT.
TFs analysis of hub genes
We further searched for the TFs of the above 9 hub genes in ChIPBase v2.0, and drew the TFs regulatory network corresponding to the hub genes, as shown in Figure 5. We can observe that FOXA1, ZBTB7A, CTCF, MYC, SP1, and CEBPB are the most common TFs that regulate the above hub genes. Among them, FOXA1 and ZBTB7A are involved in the transcriptional regulation of most hub genes, including CFAP73, LRRC74B, C12orf74, and STMND1, GPR87, TUBA4B, DNAH2.
Discussion
Fibrotic tissue is very intricately and well organized into distinct tissues, including epithelial, endothelial, stromal, and immune cells. Numerous fibrosis diseases, such as pulmonary fibrosis, arise from abnormal activation in fibroblasts, which is mainly generated by inherent normal fibroblasts in lung tissues and epithelial or endothelial cell transformation. Owing to the limitation of traditional transcriptome analysis of bulk lung tissues, the hierarchy and heterogeneity of pulmonary cells within these lineages remain unclear.
EndMT is a cellular transformation process in which endothelial cells lose their endothelial characteristics and acquire a mesenchymal cell-like phenotype (38). EndMT was originally considered to be an important biological process in embryonic development and some cardiovascular diseases (39). Recently, it has been found that EndMT is closely related to the development of fibrotic diseases. When IPF occurs, endothelial cells acquire a mesenchymal phenotype and present typical markers of myofibroblast differentiation, such as α- smooth muscle actin (α-SMA), vimentin and collagens, while reducing the expression of vascular endothelial cadherin (VE-cadherin) (40). It has been found that in the bleomycin-induced pulmonary fibrosis model, under the action of transforming growth factor-β (TGF-β) combined with Ras signal activation, endothelial cells undergo EndMT process to generate a large number of fibroblasts (41). It has also been reported that EndMT caused by heat shock protein B (small) member 1 (HSPB1) deficiency contributes to the development of pulmonary fibrosis, suggesting that HSPB1-targeted therapy may be suitable for the treatment of a range of fibrotic diseases (42). Another study showed that vildagliptin improved pulmonary fibrosis by inhibiting EndMT (43). Therefore, EndMT is considered to be one of the key factors in the occurrence of IPF. However, there is no currently relevant research that explores the relationship between EndMT and IPF from the perspective of cell heterogeneity in the fibrotic microenvironment.
Considering that epithelial-mesenchymal transition (EMT) has been widely studied in IPF (44,45), we also focused on the changes in epithelial cell scores. Interestingly, the trends in the number of epithelial cells differed between the three datasets. It has been reported that epithelial cells proliferate significantly in the early stage of IPF (46). Another study has demonstrated that in the late stage of fibrosis, the number of epithelial cells decreases, while that of fibroblasts increases (47). Therefore, we speculated that the occurrence and development of EMT might be related to the different stages of IPF. In contrast, EndMT could persist throughout IPF.
In this study, we analyzed the IPF microenvironment through single-cell sequencing data and transcriptome gene expression data. Firstly, we found that the endothelial cells scores were significantly reduced, whereas fibroblasts scores were increased in all three datasets. It was inferred that EndMT might occur in the IPF microenvironment. In order to verify our speculation, we further performed IPF microenvironment analysis using single-cell sequencing data combined with PCA, and found that the proportion of endothelial cells was significantly down-regulated, while the proportions of fibroblasts and myofibroblasts were significantly increased. Moreover, the proportion of endothelial cells decreased was similar to that of fibroblasts increase. Therefore, we speculated that EndMT might occur in the occurrence and development of IPF.
When the amount of cells changes, the function of the cells also changes considerably (48). We performed GSEA to further explore the changes in the function and phenotype of endothelial cells during EndMT. Our results indicated that BPs related to endothelial cell function, including blood vessel endothelial cell migration, endothelial cell differentiation, and negative regulation of endothelial cell proliferation, were mainly enriched in the control group. BPs related to fibroblast function, including cellular response to fibroblast growth factor stimulus and collagen fibril organization, were mainly enriched in the IPF group. This indicates that the function and phenotype of endothelial cells is also changed in the EndMT of IPF.
Gene co-expression network analysis can effectively identify functional co-expressed gene modules associated with complex diseases (34). We performed MEGENA analysis to explore key modules related to endothelial cell function and identify hub genes. We found that there were nine hub genes in the most critical modules related to endothelial cell function regulation, including CFAP73, LRRC74B, C12orf74, PTPRT, TUBA4B, STMND1, DNAH2, RSPH10B2, and GPR87. We verified the expression of these genes in the dataset and found that their expression was significantly increased in the IPF group and negatively correlated with the endothelial cell scores, indicating that these genes are likely involved in the occurrence of EndMT in IPF. The previous view that PTPRP is closely related to cell adhesion, PTPRT interacts with endothelial cadherin (E-cadherin), and this interaction leads to dephosphorylation of E-cadherin, which affects the stability of the junction complex (49). During the EndMT process, endothelial cells lost intercellular adhesion, changed the cytoskeleton organization and acquired stronger migration and invasion ability (50), indicating that PTPRP may participate in the EndMT process by affecting cell adhesion and promote the development of IPF. The G protein-coupled receptor GPR87 is overexpressed in a variety of cancers, and studies have reported that GPR87 plays a key role in the proliferation of lung cancer cells, and indicated its potential as a new target for lung cancer therapy (51-53). GPR87 overexpression promotes EMT, significantly reduces the expression of the epithelial marker E-cadherin and increases the expression of the mesenchymal cell marker N-cadherin, and enhances the invasive and metastatic phenotype of cancer cells, which is associated with poor patient prognosis (54). TUBA4B can act as a tumor suppressor in various cancers (55,56), and low expression of TUBA4B is a predictor of poor prognosis and regulated cell proliferation in non-small cell lung cancer (57). The key genes obtained from the above screening may become new potential targets for the pathogenesis of IPF, and their correlation with IPF should be explored in future studies.
We further analyzed the upstream TFs of these hub genes, and found that FOXA1 and ZBTB7A may be involved in the transcriptional regulation of most hub genes. Studies have shown that long non-coding RNA 19 attenuates lipopolysaccharide-induced pulmonary fibrosis by regulating miR-423-5p/FOXA1 (58), and demonstrated increased expression of FOXA1 in isolated lung fibrotic tissues (59). ZBTB7A has been reported to play an important role in tumor metastasis (60), and ZBTB7A promotes tumor invasion by downregulating the expression of E-cadherin (61). High expression of TGF-β can combine with E-cadherin and β-catenin to inhibit ZBTB7A (62), and Smad4, a key component of TGFβ signaling, was found to interact with ZBTB7A (63). It indicated that TFs may participate in the signal transduction of IPF by targeting hub genes.
In this study, we discussed the heterogeneity of cells in terms of the IPF microenvironment for the first time, and demonstrated that EndMT may occur throughout the IPF process.
Acknowledgments
Funding: This study was supported by grants from the National Natural Science Foundation of China (Nos. 82100061 and 82173855); the Zhejiang Province Natural Science Foundation of China (Nos. LQ21H310004, LY20H310001, and LQ21H310005); the Medical and Health Research Program of Zhejiang (Nos. 2021443880 and 2021KY019); the Chinese Medicine Research Program of Zhejiang Province (Nos. 2018ZZ006, 2020ZZ003, 2021ZQ012, 2021ZA006, 2021ZQ012, and 2022ZZ001); the “10000 Talents Plan” of Zhejiang Province to Ping Huang (No. 2020R52029); the National Natural Science Foundation of China and the Swedish Research Council Cooperative Research Project (No. 8211101233); the National Natural Science Foundation of China (No. 8217131437); and the Zhejiang Provincial Science and Technology Department’s “top soldier” and “leading wild goose” R & D project (No. 2022C03116).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1438/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1438/coif). All authors report that this study was supported by grants from the National Natural Science Foundation of China (Nos. 82100061 and 82173855); the Zhejiang Province Natural Science Foundation of China (Nos. LQ21H310004, LY20H310001, and LQ21H310005); the Medical and Health Research Program of Zhejiang (Nos. 2021443880 and 2021KY019); the Chinese Medicine Research Program of Zhejiang Province (Nos. 2018ZZ006, 2020ZZ003, 2021ZQ012, 2021ZA006, 2021ZQ012, and 2022ZZ001); the “10000 Talents Plan” of Zhejiang Province to Ping Huang (No. 2020R52029); the National Natural Science Foundation of China and the Swedish Research Council Cooperative Research Project (No. 8211101233); the National Natural Science Foundation of China (No. 8217131437); and the Zhejiang Provincial Science and Technology Department’s “top soldier” and “leading wild goose” R & D project (No. 2022C03116). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ye Z, Hu Y. TGF-β1: Gentlemanly orchestrator in idiopathic pulmonary fibrosis Int J Mol Med 2021;48:132. (Review). [Crossref] [PubMed]
- Zurkova M, Kriegova E, Kolek V, et al. Effect of pirfenidone on lung function decline and survival: 5-yr experience from a real-life IPF cohort from the Czech EMPIRE registry. Respir Res 2019;20:16. [Crossref] [PubMed]
- Raghu G, Remy-Jardin M, Myers JL, et al. Diagnosis of Idiopathic Pulmonary Fibrosis. An Official ATS/ERS/JRS/ALAT Clinical Practice Guideline. Am J Respir Crit Care Med 2018;198:e44-68. [Crossref] [PubMed]
- George PM, Patterson CM, Reed AK, et al. Lung transplantation for idiopathic pulmonary fibrosis. Lancet Respir Med 2019;7:271-82. [Crossref] [PubMed]
- Spagnolo P, Kropski JA, Jones MG, et al. Idiopathic pulmonary fibrosis: Disease mechanisms and drug development. Pharmacol Ther 2021;222:107798. [Crossref] [PubMed]
- John AE, Joseph C, Jenkins G, et al. COVID-19 and pulmonary fibrosis: A potential role for lung epithelial cells and fibroblasts. Immunol Rev 2021;302:228-40. [Crossref] [PubMed]
- Raghu G, Rochwerg B, Zhang Y, et al. An Official ATS/ERS/JRS/ALAT Clinical Practice Guideline: Treatment of Idiopathic Pulmonary Fibrosis. An Update of the 2011 Clinical Practice Guideline. Am J Respir Crit Care Med 2015;192:e3-19. [Crossref] [PubMed]
- Zhang S, Chen H, Yue D, et al. Long non-coding RNAs: Promising new targets in pulmonary fibrosis. J Gene Med 2021;23:e3318. [Crossref] [PubMed]
- Tahmasebi Birgani M, Carloni V. Tumor Microenvironment, a Paradigm in Hepatocellular Carcinoma Progression and Therapy. Int J Mol Sci 2017;18:405. [Crossref] [PubMed]
- Yeung TL, Leung CS, Yip KP, et al. Anticancer Immunotherapy by MFAP5 Blockade Inhibits Fibrosis and Enhances Chemosensitivity in Ovarian and Pancreatic Cancer. Clin Cancer Res 2019;25:6417-28. [Crossref] [PubMed]
- Reyfman PA, Walter JM, Joshi N, et al. Single-Cell Transcriptomic Analysis of Human Lung Provides Insights into the Pathobiology of Pulmonary Fibrosis. Am J Respir Crit Care Med 2019;199:1517-36. [Crossref] [PubMed]
- Habermann AC, Gutierrez AJ, Bui LT, et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci Adv 2020;6:eaba1972.
- Caja L, Dituri F, Mancarella S, et al. TGF-β and the Tissue Microenvironment: Relevance in Fibrosis and Cancer. Int J Mol Sci 2018;19:1294. [Crossref] [PubMed]
- Matsuda M, Seki E. The liver fibrosis niche: Novel insights into the interplay between fibrosis-composing mesenchymal cells, immune cells, endothelial cells, and extracellular matrix. Food Chem Toxicol 2020;143:111556. [Crossref] [PubMed]
- Ma PF, Gao CC, Yi J, et al. Cytotherapy with M1-polarized macrophages ameliorates liver fibrosis by modulating immune microenvironment in mice. J Hepatol 2017;67:770-9. [Crossref] [PubMed]
- Dong Y, Yang M, Zhang J, et al. Depletion of CD8+ T Cells Exacerbates CD4+ T Cell-Induced Monocyte-to-Fibroblast Transition in Renal Fibrosis. J Immunol 2016;196:1874-81. [Crossref] [PubMed]
- Chambers DC, Carew AM, Lukowski SW, et al. Transcriptomics and single-cell RNA-sequencing. Respirology 2019;24:29-36. [Crossref] [PubMed]
- Vukmirovic M, Kaminski N. Impact of Transcriptomics on Our Understanding of Pulmonary Fibrosis. Front Med (Lausanne) 2018;5:87. [Crossref] [PubMed]
- Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 2013;41:D991-5. [Crossref] [PubMed]
- Furusawa H, Cardwell JH, Okamoto T, et al. Chronic Hypersensitivity Pneumonitis, an Interstitial Lung Disease with Distinct Molecular Signatures. Am J Respir Crit Care Med 2020;202:1430-44. [Crossref] [PubMed]
- Schafer MJ, White TA, Iijima K, et al. Cellular senescence mediates fibrotic pulmonary disease. Nat Commun 2017;8:14532. [Crossref] [PubMed]
- Cecchini MJ, Hosein K, Howlett CJ, et al. Comprehensive gene expression profiling identifies distinct and overlapping transcriptional profiles in non-specific interstitial pneumonia and idiopathic pulmonary fibrosis. Respir Res 2018;19:153. [Crossref] [PubMed]
- Lawrence M, Huber W, Pagès H, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol 2013;9:e1003118. [Crossref] [PubMed]
- Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017;18:220. [Crossref] [PubMed]
- Stuart T, Butler A, Hoffman P, et al. Comprehensive Integration of Single-Cell Data. Cell 2019;177:1888-1902.e21. [Crossref] [PubMed]
- Wei J, Fang D, Zhou W. CCR2 and PTPRC are regulators of tumor microenvironment and potential prognostic biomarkers of lung adenocarcinoma. Ann Transl Med 2021;9:1419. [Crossref] [PubMed]
- Mohtar MA, Syafruddin SE, Nasir SN, et al. Revisiting the Roles of Pro-Metastatic EpCAM in Cancer. Biomolecules 2020;10:255. [Crossref] [PubMed]
- Lertkiatmongkol P, Liao D, Mei H, et al. Endothelial functions of platelet/endothelial cell adhesion molecule-1 (CD31). Curr Opin Hematol 2016;23:253-9. [Crossref] [PubMed]
- Hu B, Wu Z, Hergert P, et al. Regulation of myofibroblast differentiation by poly(ADP-ribose) polymerase 1. Am J Pathol 2013;182:71-83. [Crossref] [PubMed]
- Longhitano L, Tibullo D, Vicario N, et al. IGFBP-6/sonic hedgehog/TLR4 signalling axis drives bone marrow fibrotic transformation in primary myelofibrosis. Aging (Albany NY) 2021;13:25055-71. [Crossref] [PubMed]
- Ito K, Murphy D. Application of ggplot2 to Pharmacometric Graphics. CPT Pharmacometrics Syst Pharmacol 2013;2:e79. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Song WM, Zhang B. Multiscale Embedded Gene Co-expression Network Analysis. PLoS Comput Biol 2015;11:e1004574. [Crossref] [PubMed]
- Ma Q. Polarization of Immune Cells in the Pathologic Response to Inhaled Particulates. Front Immunol 2020;11:1060. [Crossref] [PubMed]
- Huaux F. Interpreting Immunoregulation in Lung Fibrosis: A New Branch of the Immune Model. Front Immunol 2021;12:690375. [Crossref] [PubMed]
- Piera-Velazquez S, Li Z, Jimenez SA. Role of endothelial-mesenchymal transition (EndoMT) in the pathogenesis of fibrotic disorders. Am J Pathol 2011;179:1074-80. [Crossref] [PubMed]
- Hulshoff MS, Xu X, Krenning G, et al. Epigenetic Regulation of Endothelial-to-Mesenchymal Transition in Chronic Heart Disease. Arterioscler Thromb Vasc Biol 2018;38:1986-96. [Crossref] [PubMed]
- Ursoli Ferreira F, Eduardo Botelho Souza L, Hassibe Thomé C, et al. Endothelial Cells Tissue-Specific Origins Affects Their Responsiveness to TGF-β2 during Endothelial-to-Mesenchymal Transition. Int J Mol Sci 2019;20:458. [Crossref] [PubMed]
- Choi SH, Nam JK, Kim BY, et al. HSPB1 Inhibits the Endothelial-to-Mesenchymal Transition to Suppress Pulmonary Fibrosis and Lung Tumorigenesis. Cancer Res 2016;76:1019-30. [Crossref] [PubMed]
- Phan THG, Paliogiannis P, Nasrallah GK, et al. Emerging cellular and molecular determinants of idiopathic pulmonary fibrosis. Cell Mol Life Sci 2021;78:2031-57. [Crossref] [PubMed]
- Hashimoto N, Phan SH, Imaizumi K, et al. Endothelial-mesenchymal transition in bleomycin-induced pulmonary fibrosis. Am J Respir Cell Mol Biol 2010;43:161-72. [Crossref] [PubMed]
- Suzuki T, Tada Y, Gladson S, et al. Vildagliptin ameliorates pulmonary fibrosis in lipopolysaccharide-induced lung injury by inhibiting endothelial-to-mesenchymal transition. Respir Res 2017;18:177. [Crossref] [PubMed]
- Tian R, Zhu Y, Yao J, et al. NLRP3 participates in the regulation of EMT in bleomycin-induced pulmonary fibrosis. Exp Cell Res 2017;357:328-34. [Crossref] [PubMed]
- Han Q, Lin L, Zhao B, et al. Inhibition of mTOR ameliorates bleomycin-induced pulmonary fibrosis by regulating epithelial-mesenchymal transition. Biochem Biophys Res Commun 2018;500:839-45. [Crossref] [PubMed]
- Königshoff M. Lung cancer in pulmonary fibrosis: tales of epithelial cell plasticity. Respiration 2011;81:353-8. [Crossref] [PubMed]
- Zanoni M, Cortesi M, Zamagni A, et al. The Role of Mesenchymal Stem Cells in Radiation-Induced Lung Fibrosis. Int J Mol Sci 2019;20:3876. [Crossref] [PubMed]
- Zhou Y, Zhu J, Zou L, et al. Changes in number and biological function of endothelial progenitor cells in hypertension disorder complicating pregnancy. J Huazhong Univ Sci Technolog Med Sci 2008;28:670-3. [Crossref] [PubMed]
- Scott A, Wang Z. Tumour suppressor function of protein tyrosine phosphatase receptor-T. Biosci Rep 2011;31:303-7. [Crossref] [PubMed]
- Islam S, Boström KI, Di Carlo D, et al. The Mechanobiology of Endothelial-to-Mesenchymal Transition in Cardiovascular Disease. Front Physiol 2021;12:734215. [Crossref] [PubMed]
- Kita Y, Go T, Nakashima N, et al. Inhibition of Cell-surface Molecular GPR87 With GPR87-suppressing Adenoviral Vector Disturb Tumor Proliferation in Lung Cancer Cells. Anticancer Res 2020;40:733-41. [Crossref] [PubMed]
- Yasui H, Nishinaga Y, Taki S, et al. Near-infrared photoimmunotherapy targeting GPR87: Development of a humanised anti-GPR87 mAb and therapeutic efficacy on a lung cancer mouse model. EBioMedicine 2021;67:103372. [Crossref] [PubMed]
- Gugger M, White R, Song S, et al. GPR87 is an overexpressed G-protein coupled receptor in squamous cell carcinoma of the lung. Dis Markers 2008;24:41-50. [Crossref] [PubMed]
- Ahn HM, Choi EY, Kim YJ. GPR87 Promotes Metastasis through the AKT-eNOS-NO Axis in Lung Adenocarcinoma. Cancers (Basel) 2021;14:19. [Crossref] [PubMed]
- Guo J, Li Y, Duan H, et al. LncRNA TUBA4B functions as a competitive endogenous RNA to inhibit gastric cancer progression by elevating PTEN via sponging miR-214 and miR-216a/b. Cancer Cell Int 2019;19:156. [Crossref] [PubMed]
- Zhou YG, Sun F, Zhou YF. Low expression of lncRNA TUBA4B promotes proliferation and inhibits apoptosis of colorectal cancer cells via regulating P15 and P16 expressions. Eur Rev Med Pharmacol Sci 2020;24:3023-9. [PubMed]
- Chen J, Hu L, Wang J, et al. Low Expression LncRNA TUBA4B is a Poor Predictor of Prognosis and Regulates Cell Proliferation in Non-Small Cell Lung Cancer. Pathol Oncol Res 2017;23:265-70. [Crossref] [PubMed]
- Mu X, Wang H, Li H. Silencing of long noncoding RNA H19 alleviates pulmonary injury, inflammation, and fibrosis of acute respiratory distress syndrome through regulating the microRNA-423-5p/FOXA1 axis. Exp Lung Res 2021;47:183-97. [Crossref] [PubMed]
- Hanmandlu A, Zhu L, Mertens TCJ, et al. Transcriptomic and Epigenetic Profiling of Fibroblasts in Idiopathic Pulmonary Fibrosis. Am J Respir Cell Mol Biol 2022;66:53-63. [Crossref] [PubMed]
- Singh AK, Verma S, Kushwaha PP, et al. Role of ZBTB7A zinc finger in tumorigenesis and metastasis. Mol Biol Rep 2021;48:4703-19. [Crossref] [PubMed]
- Guo C, Zhu K, Sun W, et al. The effect of Pokemon on bladder cancer epithelial-mesenchymal transition. Biochem Biophys Res Commun 2014;443:1226-31. [Crossref] [PubMed]
- Li W, Kidiyoor A, Hu Y, et al. Evaluation of transforming growth factor-β1 suppress Pokemon/epithelial-mesenchymal transition expression in human bladder cancer cells. Tumour Biol 2015;36:1155-62. [Crossref] [PubMed]
- Yang Y, Cui J, Xue F, et al. Pokemon (FBI-1) interacts with Smad4 to repress TGF-β-induced transcriptional responses. Biochim Biophys Acta 2015;1849:270-81. [Crossref] [PubMed]
(English Language Editor: A. Kassem)