Immuno-regulated common markers but different network signatures in two associated cancers: evidences from epigenetic treatment
Introduction
The retinoblastoma (RB) susceptibility gene RB1 is known to play a causal role in human cancer (1-3). In particular, the gene inactivation in chromosome band 13q14 has revealed RB formation, an identification which was derived from specific mutations. The RB1 gene and its protein explain key mechanisms of oncogenesis, development, and gene regulation, and help our understanding of genetic diagnosis of susceptibility for other RB1-dysfunction driven cancers, such as the human osteosarcoma (HOS) (4). In the past, a few hereditary HOS risk factors have been identified, but these rare events account for relatively few patients. It is also well-known, however, that survivors of the inherited form of RB are at an increased risk to develop second primary HOS compared to the general population or to survivors of non-inherited RB (5).
Cancer is considered to be a genetic disease, but other types of alterations are exerting influences. For instance, epigenetic modifications may regulate gene expression and, thus, cancer development. Epigenetic dysregulation may precede other transforming events, ranging from focal mutations to genome-wide instability (6,7). Especially recent results have emphasized the role of ‘methylomes’ (i.e., genome-scale methylation maps obtained through high-throughput methods) in establishing cancer hallmarks (8). Among the several types of known epigenetic modifications, DNA methylation is of particular clinical interest due to its association with cancer initiation and progression, and its role as predictive biomarker for response to chemotherapy (9). Linking epigenetic changes with drug resistance may form a basis for the design of future treatments strategies combining epigenetic and chemotherapeutic regimens.
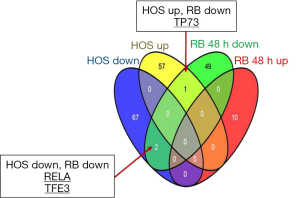
We provide comparative evidence derived from the analyses of RB and HOS cells treated with a demethylating agent (DAC). In particular, the objective if the study is to analyze the marker role of genes that were detected as differentially expressed (DEG) in both cell types, and assess the network signatures induced by these common genes by targeting both co-expression associative dynamics and interactions between their products. In earlier work (10,11), we focused on the osteosarcoma-derived HosDXR150 cell line and found that its proliferation was effectively reduced by treatment with the DAC 5-Aza-dC (decitabine) alone, among other types of inhibitors. We also obtained a DEG profile from time course microarray experiments on the RB-derived WERI-RB1 cell line treated with 5-Aza-dC only. The DEG lists and their annotations are provided in Tables 1,2, considering both cancer profiles measured 48 h after DAC treatment. A Venn diagram is shown in Figure 1 for comparative evaluations restricted to common DEGs, either up- or down-regulated.

Full table

Full table
Methods
We briefly summarize the data generation aspects relevant to the novel developments, and refer the readers for further details on HOS and RB treatments to our previous work (10,11). Here, we embrace networks for inference purposes, and following known techniques (12-14).
HosDXR150 cells were treated with 2.5 μM 5-Aza-2’-deoxycytidine (5-Aza-dC) for an incubation period of 24-96 h, in absence (control) or presence of drugs. Total RNA was isolated from treated and untreated cells after 48 h using TRIZOL reagent (Invitrogen). Subsequent cDNA microarray expression analysis was performed by using MWG Hybridization Service (MWG Biotech AG). For each experimental point 10 μg of total RNA from the control (reference pool) and the sample (test pool) were labeled with Cy3 and Cy5 respectively. Each channel was scanned three times with increasing photomultiplier gain settings using Scanner 418/428 equipment (Affimetrix) at 10 µm resolution, thereby ensuring coverage of the full dynamic range. The ImaGene intensity values were processed using the MAVI software package (MWG Biotech AG).
DEGs were selected by fixing 1.5-fold change (up-regulated if ≥1.5 and down-regulated if ≤−1.5) in log2 expression ratios, statistically significant at a cutoff P value of 0.01 (Wilcoxon test). Benjamini-Hochberg correction for multiple testing was applied. For GO term analysis, biological process annotations were obtained from the package GO database (v.2.9.0), whereas for pathway enrichment analysis the ClueGO software package (2.0.6) was used (http://apps.cytoscape.org/apps/cluego) (15). The statistical test used for enrichment was the right-sided hypergeometric test, and only terms with P values <0.05 and at least three genes per term were selected, followed by a multiple testing correction using the Benjamini-Hochberg method. The pathway sources were KEGG, REACTOME and WikiPathways.
The RB-derived cell line Weri-RB1, obtained from ATCC (Rockinville, MD), was treated with 5-Aza-dC (versus untreated control) in a time course experiment measuring gene expression profiles at 3 specific time points after treatment initiation, i.e., after 48, 72 and 96 h. Total RNA samples were isolated using TRIZOL (Invitrogen). After cDNA conversion, microarray expression analysis was carried out using PIQORTM Cell Death Human Sense Microarrays (Miltenyi Biotech) containing 200-mer oligo-probes covering 494 human genes. Mean signal and local background intensities were obtained for each spot on the microarray images using the ImaGeneTM software package (BioDiscovery).
Assessment of gene expression profiles at the three time points was achieved by using a Cy5/Cy3 customized platform containing about 500 genes related to apoptosis, cell death and inflammation. Local background was subtracted from the signal to obtain the signal intensity, after which the Cy5/Cy3 ratio was calculated. Subsequently, the mean of the ratios of 4 spots for the same gene was computed. The ratios were normalized using the Median and the Lowess methods. Quality filtering was applied for the calculation of the Cy5/Cy3 ratio to only spots/genes with at least in one channel a signal intensity 2-fold higher than the mean background, for then selecting up- and down-regulated genes.
GeneMania (http://www.genemania.org/) (16,17) was used to generate the networks, showing co-expression dynamics among the connected genes through the available Co-expression, Co-localization, Genetic interactions, Pathway, Physical interactions, Predicted and Shared protein domains (default settings). The results were imported into Cytoscape for display. GeneMania allowed an assessment of the interactions occurring at co-expression level for the DEG sets in all the measured time course profiles. The network configurations were built from the log2 expression ratio values.
Functional enrichment was obtained with David (http://david.abcc.ncifcrf.gov/) and PantherDB (http://www.pantherdb.org/) tools for GO enrichment and pathway mapping, respectively. Pathway analysis was performed using ClueGo in Cytoscape 3.1, and by selecting as annotation sources Wikipathways, KEGG and Reactome. The statistical analysis included right-sided hypergeometric test for enrichment by Benjamini-Hochberg P value correction.
Results
GO (biological processes) and pathway annotations referred to the evidences obtained from the RB-derived cell line Weri-RB1 are listed in Tables 1,2, respectively. The DEG sets enrich terms according to FDR-corrected P values. Pathway annotations for the HOS-derived cell line HosDXR150 are listed in Table 3 (up-regulated DEGs) and in Table 4 (down-regulated DEGs), respectively. We found only 3 DEG markers that were shared between the two cancer types; one of them (TP73) exhibiting a discordant regulation sign (Figure 1). From these genes as the network seed nodes, co-expression network links (dashed lines) and protein-protein interaction networks (straight lines) were built. In the figures, the blue rectangular nodes indicate DEGs, while the grey ovals represent interactors that were added to complete the connectivity patterns.
Full table

Full table
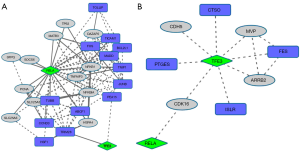
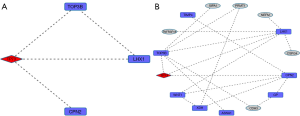
A neighbor-1 expanded network (i.e., starting from a seed node, and including the direct links with surrounding nodes) is provided for TP73 in both cancer types, due to the particular relevance of this gene. Only closest neighbor exploration of the seed gene was considered, to avoid redundant networks. The genes of interest are shown in Figure 1. Firstly, transcription factor binding to IGHM enhancer 3 (TFE3), which plays a role in immunity (activation of CD40L in T-cells) and is also known as a fusion gene associated with chromosomal translocations. Secondly, V-Rel avian reticuloendotheliosis viral oncogene homolog (RELA), deeply involved in NFkB1 signaling, and related to the PI3k-Akt cascade, inflammation, immunity, differentiations, cellular growth and tumorigenic processes. Finally, TP73 is a transcription factor participating in apoptotic response to DNA damage and essential for expressing cytokines in T-cells. It appears that the only commonly and DEG markers bring an immune system signature.
Continuing with the annotations, the TFE3 encoded protein promotes the expression of genes downstream of TGF-β signaling. It activates the expression of CD40L in T-cells, thereby playing a role in T-cell-dependent antibody responses in activated CD4-positive T-cells and thymus-dependent humoral immunity. NFkB is a pleiotropic transcription factor ubiquitously involved in several biological processes, and encompassing NFkB1 and NFkB2 bound to either RELA or RELB. The activity of NFkB is also affected by various mechanisms of post-translational modification and sub-cellular compartmentalization, as well as by interactions with other co-factors (18).
The jointly RELA- and TFE3-induced network is denser in RB cells than in HOS ones (Figure 2). In particular, the RELA sub-networks are populated with DEGs. An interesting PIN link for TFE3 is TRIM28. The latter gene encodes a protein that is localized in the nucleus, where it mediates transcriptional control by interacting with the KRAB (Kruppel-associated box) repression domain present in many transcription factors. According to the Human Gene Databse annotation tool (http://www.genecards.org/) TRIM28 is associated with specific chromatin regions, and mediates epigenetic gene silencing by recruiting CHD3, a subunit of the nucleosome remodeling and deacetylation (NuRD) complex, and SETDB1 to the promoter regions of KRAB target genes. Also, it enhances transcriptional repression via increases in H3 Lys-9 methylation, and decreases in histone H3 Lys-9 and Lys-14 acetylation, and also coordinates the disposition of HP1 proteins. Finally, it acts as an inhibitor of E2F1 activity by stimulating E2F1-HDAC1 (histone de-acetylase-1) complex formation and inhibiting E2F1 acetylation. Interestingly, it participates in E2F1-mediated apoptosis in the absence of RB1, and mediates the transcriptional repressing activity of FOXP3 and the suppressive function of regulatory T-cells (Treg) (19). In HOS cells, the two target genes induce a much smaller network (only one co-expressed gene appears). A role in immunity is appearing but remains to be deciphered, in view of the evidences from the two gene markers (RELA and TFE3).
TP73 encodes a member of the p53 family of transcription factors involved in cellular responses to stress and development. It maps to a region on chromosome 1p36 that is frequently deleted in tumors, and that is thought to contain multiple tumor suppressor genes. It participates in the apoptotic response to DNA damage, possibly as a tumor suppressor protein. Down-regulated TP73 in RB cells (Figure 3) induces a small sub-network, which is expanded when neighbor-1 (indirect) interactors are linked. Among the DEGs that interact with the target, DAXX appears relevant. This gene encodes a multifunctional protein that interacts with a wide range of proteins, including apoptosis-related FAS. The indirect links with DEGS that are reached through the neighbor-1 links are, in general, functionally relevant.
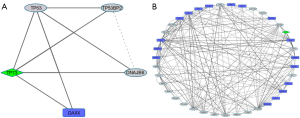
In HOS cells, TP73 has a discordant sign (up-regulation) and the induced sub-network is entirely co-expressed. Oncogenic WNT1 is one of the uncovered indirect associations, as also TIMP4 which encodes an inhibitor of matrix metalloproteinases (MMPs), a group of peptidases that is involved in degradation of the extracellular matrix (Figure 4).
Conclusions
Three DEGs were identified as shared markers in two cell lines derived from associated cancers after treatment with the same DAC. They were analyzed alone and through their induced co-expression and protein-protein interaction network dynamics. Exploiting the concept of networks and their modularity offers several advantages regarding the prediction of regulatory programs occurring at a gene ensemble scale rather than single genes.
Apart from the different sub-network configurations that are necessarily gene-specific, what is relevant from our analysis is the emergence of differential signatures for markers of two associated cancers (RB and HOS) that were both detected as DEG after DAC treatment. The evidences here presented conform with our findings from previous studies, especially indicating that ensemble network markers can provide more effective disease signatures than single genes in HOS cells.
While establishing new cancer phenotypes through networks remains a valid possibility, testing is quite clearly needed on a case-by-case basis. In the present study, for instance, it was found that with three DEG markers in common between the associated cancers and considered as potentially relevant disease phenotypes, only for two of them (RELA and TFE3), and prevalently for one cancer (RB), we could find an informative network signature, according to standard annotations. Indeed, we were able to find marks of immunity-related functions and epigenetic effects.
For the TP73 gene marker, the induced networks have shown different configurations, with the important marks of apoptosis remaining more evident in just one cancer type (RB). Both direct and indirect connectivity patterns were found to be different across the two cancers. Even after expanding the marker outreach to include more relationships across the networks compared to before, the effects did not appear to lead to homogeneous signatures. This observation was contingent in both cancers, i.e., only the closest gene and protein interactors were explored by taking advantage of the information provided by the computational tool. Further analyses could be employed with different outcomes.
The main conclusion from the examples here provided is that, despite evidence on disease phenotypes from three shared markers arising from similar experiments centered on epigenetic (DAC-driven) treatment of two associated cancers (RB and HOS), a more complete interpretation of cancer associations and/or specific versus differential role of the immune system remains hard to achieve once contextual analyses are carried out for such markers over their induced networks signatures.
Apart from deciphering the epigenetic influences exerted on the DEG cancer patterns, one limitation of this study is that the expression profiles were obtained from microarray experiments, which are known to be of minor depth and accuracy compared to comprehensive high-throughput settings, such as RNA-Seq. Another limitation is that other types of regulation may underlie the DAC-driven phenotypes induced in the two examined cell lines, but for these possible causal influences data are not available at this time.
In order to overcome such limitations, a current direction of work is to move from cell lines to patient samples, especially with HOS, and run RNA-Seq experiments to generate data. Finally, the two tumors offer relatively limited coverage, compared to other cancers, thus need additional in-depth analyses, we are confidently looking into the direction of research named ‘integrative omics’ to provide further applications and novel insights in different primary cancers and treatment scenarios.
In conclusion, network medicine can elucidate cancer aspects by performing integrative multiscale inference on identified oncoepigenetic network signatures potentially linked to disease hallmarks. However, it emerges from the performed comparative analysis that DEG genes after epigenetic treatment build only isolated hotspots rather than cancer signatures, and therefore it would be wiser to explore differential cancer molecular profiles as those uncovered by network modules.
Acknowledgements
The author thanks C Cinti for discussions on the two cancers and for generating the data that were published in the reported earlier work.
Footnote
Conflicts of Interest: The author has no conflicts of interest to declare.
References
- Lee WH, Bookstein R, Lee EY. Studies on the human retinoblastoma susceptibility gene. J Cell Biochem 1988;38:213-27. [Crossref] [PubMed]
- Issing WJ, Wustrow TP, Oeckler R, et al. An association of the RB gene with osteosarcoma: molecular genetic evaluation of a case of hereditary retinoblastoma. Eur Arch Otorhinolaryngol 1993;250:277-80. [Crossref] [PubMed]
- Khosravi A, Shahrabi S, Shahjahani M, et al. The bone marrow metastasis niche in retinoblastoma. Cell Oncol (Dordr) 2015;38:253-63. [Crossref] [PubMed]
- Hansen MF, Koufos A, Gallie BL, et al. Osteosarcoma and retinoblastoma: a shared chromosomal mechanism revealing recessive predisposition. Proc Natl Acad Sci U S A 1985;82:6216-20. [Crossref] [PubMed]
- Chauveinc L, Mosseri V, Quintana E, et al. Osteosarcoma following retinoblastoma: age at onset and latency period. Ophthalmic Genet 2001;22:77-88. [Crossref] [PubMed]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012;13:484-92. [Crossref] [PubMed]
- Rodríguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med 2011;17:330-9. [Crossref] [PubMed]
- Witte T, Plass C, Gerhauser C. Pan-cancer patterns of DNA methylation. Genome Med 2014;6:66. [Crossref] [PubMed]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 2011;11:726-34. [Crossref] [PubMed]
- Capobianco E, Mora A, La Sala D, et al. Separate and combined effects of DNMT and HDAC inhibitors in treating human multi-drug resistant osteosarcoma HosDXR150 cell line. PLoS One 2014;9:e95596. [Crossref] [PubMed]
- Malusa F, Taranta M, Zaki N, et al. Time-course gene profiling and networks in demethylated retinoblastoma cell line. Oncotarget 2015;6:23688-707. [Crossref] [PubMed]
- Chen L, Liu R, Liu ZP, et al. Detecting early-warning signals for sudden deterioration of complex diseases by dynamical network biomarkers. Sci Rep 2012;2:342. [Crossref] [PubMed]
- Deng S, Qi J, Stephen M, et al. Network-based identification of reliable bio-markers for cancers. J Theor Biol 2015;383:20-7. [Crossref] [PubMed]
- Mora A, Taranta M, Zaki N, et al. Ensemble inference by integrative cancer networks. Front Genet 2014;5:59. [Crossref] [PubMed]
- Bindea G, Mlecnik B, Hackl H, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009;25:1091-3. [Crossref] [PubMed]
- Mostafavi S, Ray D, Warde-Farley D, et al. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol 2008;9 Suppl 1:S4. [Crossref] [PubMed]
- Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010;38:W214-20. [Crossref] [PubMed]
- Vazquez-Santillan K, Melendez-Zajgla J, Jimenez-Hernandez L, et al. NF-κB signaling in cancer stem cells: a promising therapeutic target? Cell Oncol (Dordr) 2015;38:327-39. [Crossref] [PubMed]
- Ohkura N, Kitagawa Y, Sakaguchi S. Development and maintenance of regulatory T cells. Immunity 2013;38:414-23. [Crossref] [PubMed]


