Has MET met its match?
The last decade has seen a revolution in both our understanding of the genomic landscape of a wide range of malignancies and the recognition that a spectrum of tumors harbors dominant oncogenic alterations leading to a unique dependence of these tumors to the presence/activity of the oncogene, called oncogene addiction. In tumors where the activity of such oncogenic gene products can be successfully inhibited pharmacologically, responses can be dramatic. Leading examples of such aberrations include BCR-ABL positive chronic myeloid leukemia, KIT-mutated gastrointestinal stromal tumor (GIST) sarcomas and epidermal growth factor receptor (EGFR)-mutated lung adenocarcinomas, and in patients with these tumors, the treatment landscape has been completely redrawn. With the advent of high-throughput genomics technology and through major international collaborative efforts, such as The Cancer Genome Atlas (TCGA), comprehensive information is now available of genomic alterations in many common and less common malignancies leading some to question whether the information to be learned about highly actionable alterations is reaching saturation. It is with this background that we will review recent major findings, including the manuscript of Frampton et al. (1), leading to a breakthrough in our understanding of the oncogenic role and long-proposed but elusive actionability of the MET tyrosine kinase.
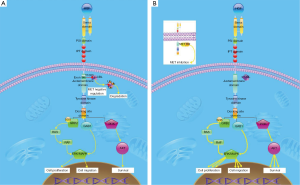
The MET gene encodes for the hepatocyte growth factor receptor (2), a receptor tyrosine kinase whose sole ligand is the hepatocyte growth factor (HGF). It was identified in 1984 from a chemically transformed osteosarcoma cell line (3). Its sequence is distinct from the ERBB family of receptor tyrosine kinases, but has most similarity to insulin receptor and the oncogene ABL (4). A critical role in cellular mobility was identified during brain development (5) with important functions in embryogenesis, wound healing and branching morphogenesis. MET activation of downstream signaling pathways are diverse, and in some cases additive. The docking region can bind GAB1 and GRB2 which leads to complex formation activating MAP-kinase and PI3K pathways. These signals can be cooperatively enhanced by adaptor molecules provided by integrins. In addition, SEMA ligands can bind plexin receptors, and plexin-MET interaction can result in MET activation in the absence of HGF. Interactions between CD44 and MET are also important leading to activation of the RAS pathway via SOS (6). The activation of the MAPK pathway via MEK1 and p38 has proliferative and anti-apoptotic roles critical for oncogenesis. Phosphatidylinositol-3-kinase (PI3K) activation also leads to an anti-apoptotic effect, as well as a pro-proliferative effect. When these activations are unopposed or constitutive, their effect is oncogenic.
MET internalization occurs in pathways of both activation and inhibition. However, the trafficking of these endosomes to lysosomes is critical to pathway inactivation. CBL is an E3 ubiquitin protein ligase recruited upon the phosphorylation of a tyrosine at position 1003 in the juxtamembrane domain of MET. This results in multiple ubiquitylation, causing fusion with multivesicular bodies which prevent membrane recycling, and ultimately targets the protein for lysosomal degradation. The balance between membrane recycling and targeting for lysosomal fusion maintains or terminates MET signaling.
The MET gene maps to chromosome 7, at cytogenetics location 7q31.2 (7) which constitutes 7:116,672,404–116,798,385 (8) in the UCSC genome browser. Human tumors exhibit a variety of alterations in MET. Chromosomal aberrations that involve this region range from chromosomal aneuploidies including trisomies (9) and tetrasomies at the lower level of copy number increase to high polysomies and high level amplifications. Mutational events have also been demonstrated, some of which are somatic and some that are germline. MET mutations were identified in patients with hereditary papillary renal carcinoma (10), including H1112R point mutations, with additional point mutations in M1149T, V1206L,V1238I, D1246N, Y1248C, and L1213V reported (11), all of which flank the tyrosine kinase domain. Tyrosine kinase domain mutations, however, are not commonly seen in lung cancer where rare missense mutations are seen in multiple domains, including mutations of residue Y1003 specifically affecting the CBL binding site. Rare chromosomal fusions of MET have also been reported, such as KIF5B-MET in lung adenocarcinoma and TFG-MET fusion protein in thyroid papillary carcinoma.
In contrast, the overexpression of MET as measured by IHC is found in a large proportion of lung cancers (12,13), and increased HGF expression is common (12). In one series, 72% of cases were MET positive with 13% of cases showing strong IHC reactivity. Expression of p-MET is seen in 73% of cases with varying intensity. Amplification is reported in 1.3–2.2% (14,15) but overall CNA including high polysomy in 12% (16). It is notable that heterogeneity within tumors is observed (17). There is inconsistent concordance between IHC and FISH for copy number (18,19); while there is some relationship between staining and amplification, IHC is positive in a much larger proportion of cases than those that ultimately are amplified. These observations have resulted in significant hurdles in development of the optimal biomarker for MET targeting. MET gene amplification is an important mechanism of acquired resistance in EGFR mutated lung adenocarcinomas treated with EGFR targeting tyrosine kinase inhibitors (20). This occurs in a proportion of tumors (5–20%), and the mechanism of resistance was linked to activation of ERBB3 and the PI3K pathways, effectively bypassing the EGFR blockade (21) with recent findings suggestive of this occurring as a result of uncoupling of EGFR oncogenic activity from its kinase function (22). This leads to another scenario in which MET targeting may be of clinical significance.
Despite these molecular alterations leading to increased signaling with oncogenic effect, inhibition of this pathway has not been consistently successful. Onartuzumab (MetMab) is a monovalent humanized monoclonal antibody. In a phase I study, the combination of erlotinib and onartuzumab was tolerated, and one response was seen in an adenocarcinoma with 3+ MET positive IHC. A randomized phase II study of onartuzumab with erlotinib (23) was overall a negative study, but the MET IHC positive population appeared to benefit with improved PFS and OS. These encouraging findings for the MET-positive subset led to a pivotal phase III study focusing only on (24) IHC MET positive (2 or 3+ in ≥50% of tumor cells) patients. However, in a major setback for the field, this study was terminated prematurely as it did not confirm the benefit of this drug combination. A small molecule, non-ATP competitive inhibitor of MET, tivantinib, was also examined in combination with erlotinib in previously treated advanced non-squamous non-small cell lung cancer patients in a phase III study (MARQUEE study) enriching for MET-high patients following a randomized phase II study showing trends towards benefit in MET-positive, non-squamous patients. While there was a suggestion of improved PFS (but not OS, the study’s primary endpoint), the study was terminated as a result of increased incidence of interstitial lung disease and projected futility at a pre-planned analysis. More encouragingly, in one small series partial responses to the dual ALK/MET inhibitor, crizotinib were seen in patients with advanced non-small cell lung cancer (NSCLC) harboring MET genomic amplification with a higher likelihood of response in tumors with high level amplification (ratio ≥5; 67% response rate), a result which requires confirmation in a larger series (25), however has impacted clinical practice and is included in treatment guidelines, such as National Comprehensive Cancer Network (NCCN). Analogous significant responses have been reported in MET-amplified gastric adenocarcinoma patients as well in several case reports and case series with crizotinib (26) and AMG-377 (27), while several other large studies using anti-MET antibodies such as onartuzumab and rilotumumab focusing on MET IHC positive patients were terminated prematurely due to futility similar to the above cited lung studies. Overall, the above results have questioned the utility of MET inhibition in patients selected by MET expression and raised the possibility that smaller subsets of biomarker-enriched patients based on gene level alterations on the other hand might benefit more significantly from MET inhibitor therapy, specifically MET tyrosine kinase inhibitor therapy.
While MET mutations in the tyrosine kinase domain in lung cancer are not common, a unique mechanism of MET activation via mutational events affecting the RNA splice acceptor or donor sites of exon 14 of MET was first reported in 2006. Such mutations that result in an in-frame deletion of the juxtamembrane domain (termed exon 14 skipping) result in loss of tyrosine 1003, the CBL ubiquitin ligase site, with resulting decreased MET ubiquitylation (Figure 1) (28). The increased MET levels and prolonged activation result in downstream MAPK activation and might be one mechanism of oncogenicity, while increased kinase activation due to conformational changes might be another. The activity can be blocked by small molecule MET inhibitors. Later, Onozato et al. (29) found MET amplification in 1.4% of lung ADCA and exon 14 skipping mutations in 3.3%. Subsequently MET exon14 skipping mutations have been found in about 4% of lung adenocarcinoma in larger series, including TCGA (14,15,30). In these series, MET exon 14 skipping mutations were mutually exclusive with oncogenic driver mutations in EGFR, KRAS, ALK and BRAF in line with this genetic event as an early driver mutational event.
Given the ready availability of MET inhibitors, such as crizotinib, several groups have attempted therapy in patients with MET exon 14 skipping. Using crizotinib, cabozantinib, or capmatinib (INC280), Frampton et al. reported partial responses in two patients with lung cancer (large cell and squamous), Paik et al. (30) in 4 patients with lung adenocarcinoma, Liu et al. (31) in one patient with sarcomatoid carcinoma, and three individual additional reports of responses in lung cancer (2 AdCa and 1 SqCa) (32-34) have also been noted.
In the stellar manuscript by Frampton et al., 38,028 tumor specimens from a period of 2012 to February 2015 were analyzed using the FoundationOne test, a next generation sequencing panel. Alterations that potentially affected MET exon 14 splicing were identified in 221 cases (0.6% of cases overall), including 131 lung adenocarcinomas (for an overall frequency of 3% in this subtype), 62 other lung carcinomas and 15 tumors of unknown primary, 6 gliomas and 7 in miscellaneous tumor types. A very wide array of 126 distinct sequence variants were noted strongly arguing against more limited exon sequencing for the detection of MET genomic abnormalities. Intriguingly, six cases with MET exon 14 skipping were sarcomatoid carcinomas, a rare subtype of lung carcinoma; in another recent manuscript Liu et al. described these mutations in over 20% of this highly aggressive and treatment-refractory subtype, immediately expanding the horizon for the molecular testing and therapy for patients with this most devastating type of lung cancer. Notably, these complimentary observations make biological sense given the role of MET kinase in mesenchymal transformation/EMT as well as increased invasion/metastatic capacity. The majority of lung tumors in the other carcinoma group were in a NSCLC NOS category. This group also examined the oncogenic effect of MET exon 14 deletion in human cells, MET exon 15 deletions in mouse cells, and the response of these cells to capmatinib. Among the 221 patients, 3 were treated with MET inhibitors, and they each experienced a partial response—a chest wall histiocytic sarcoma patient treated with crizotinib, and two NSCLC patients (a large cell and poorly differentiated squamous carcinoma) treated with capmatinib, an investigational MET inhibitor. Further complicating this area are the facts that MET exon 14 skipping alterations overlap with MET amplification as well as can concurrently occur with other mutations, such as PI3-kinase mutations that can potentially limit MET inhibitor efficacy. Lastly, the manuscript of Frampton et al. also highlighted the strong concurrence of MET exon 14 skipping aberrations with copy number changes in MDM2 and CDK4.
The exciting and pivotal findings of Frampton et al. in the background of the collection of contemporaneous publications defining the frequency and actionability of MET exon 14 skipping mutations suddenly bring clarity to a decade-long quest for defining subsets of patients for MET targeting. While the number of patients reported overall remains low, the very high response rate and the quality of the responses call imminently for the incorporation of MET testing in the management of advanced NSCLC. Patients identified with such abnormalities should be candidates for ongoing clinical studies focusing on the evaluation of MET inhibitors or should be offered therapy with commercially available MET inhibitors, such as crizotinib or cabozantinib through the course of their illness. The frequency of MET exon 14 skipping alterations in the range of 2–4% of lung adenocarcinoma is in line with frequencies of actionable ALK alterations. Therefore, testing for such mutations in a comprehensive fashion should be immediately incorporated into multi-gene testing panels. Several ongoing studies, including studies with the highly potent and selective MET inhibitors, INC280, MGCD265 and MGCD516 now focus on careful molecular selection for the inclusion of patients and should be strongly supported for better understanding of the value of MET inhibition (Table 1).

Full table
Despite these exciting observations and the emergence of exon 14 skipped MET-positive lung cancers as a unique entity, a large number of open questions remain to be answered in future investigations. Efforts need to be expanded to identify optimal biomarkers for selection of these cases. Given the broad spectrum of mutations affecting the 5’ and 3’ splice sites, next generation sequencing platforms appear to be in the lead; however, the unique protein product generated opens avenues for antibody development to be used in IHC-based assays. Exciting technological developments potentially allow the detection of these unique genetic alterations from ctDNA, facilitating the conduct of ongoing and future clinical studies. In addition, further efforts will need to be undertaken as to the actual mechanisms of oncogenicity, impact on CBL-mediated degradation, specific contributions of MET kinase activity to EMT/sarcomatoid transformation, and invasive/metastatic capacity. In particular, of high priority is to understand whether MET exon 14 skipping mutations might preferentially affect invasive/EMT-type subclones of lung cancers. Also, of immediate value will be in vitro as well as tissue-based studies to model/assess mechanisms of acquired drug resistance such as through secondary MET mutations or bypass pathway activation. In addition to supporting ongoing studies of MET inhibitors, it is critically important to model and develop combination strategies as single-agent MET inhibition is anticipated to be of short-term value. Future efforts will need to be focused on developing combination strategies with other targeted and immunotherapeutic agents to extend the benefits of such treatment interventions where a very high initial response rate is anticipated based on the available reports but the development of resistance might limit long-term impact on outcomes.
All in all, last year clearly put MET exon 14 skipping mutations on the map as a key actionable lung cancer oncogene with significant frequency to warrant incorporation into routine testing algorithms in order to be able to offer patients with advanced MET exon 14 skipped lung cancers an additional line of highly active treatment option. Finally, it does seem that after 20 years of active research of this intriguing pathway, MET has “met its match”. It is up to translational medicine to follow the lead and extend the benefits quickly and broadly to those of our patients harboring malignancies driven by oncogenic MET signaling.
Acknowledgements
None.
Footnote
Provenance: This is a Guest Editorial commissioned by Section Editor Ying Liang, MD, PhD [Associate Professor, Department of Medical Oncology, Sun Yat-sen University Cancer Center (SYSUCC), Guangzhou, China].
Conflicts of Interest: B Halmos: clinical research funding from Mirati. The other authors have no conflicts of interest to declare.
References
- Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov 2015;5:850-9. [Crossref] [PubMed]
- Bottaro DP, Rubin JS, Faletto DL, et al. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science 1991;251:802-4. [Crossref] [PubMed]
- Cooper CS, Park M, Blair DG, et al. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature 1984;311:29-33. [Crossref] [PubMed]
- Dean M, Park M, Le Beau MM, et al. The human met oncogene is related to the tyrosine kinase oncogenes. Nature 1985;318:385-8. [Crossref] [PubMed]
- Powell EM, Mars WM, Levitt P, et al. Hepatocyte growth factor/scatter factor is a motogen for interneurons migrating from the ventral to dorsal telencephalon. Neuron 2001;30:79-89. [Crossref] [PubMed]
- Orian-Rousseau V, Chen L, Sleeman JP, et al. CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes Dev 2002;16:3074-86. [Crossref] [PubMed]
- Available online: http://www.omim.org/geneMap/7/519?start=-3&limit=10&highlight=519
- Available online: http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg38&position=chr7:116672404-116798385&dgv=pack&knownGene=pack&omimGene=pack
- Zhuang Z, Park WS, Pack S, et al. Trisomy 7-harbouring non-random duplication of the mutant MET allele in hereditary papillary renal carcinomas. Nat Genet 1998;20:66-9. [Crossref] [PubMed]
- Schmidt L, Junker K, Weirich G, et al. Two North American families with hereditary papillary renal carcinoma and identical novel mutations in the MET proto-oncogene. Cancer Res 1998;58:1719-22. [Crossref] [PubMed]
- Schmidt L, Duh FM, Chen F, et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet 1997;16:68-73. [Crossref] [PubMed]
- Ma PC, Tretiakova MS, MacKinnon AC, et al. Expression and mutational analysis of MET in human solid cancers. Genes Chromosomes Cancer 2008;47:1025-37. [Crossref] [PubMed]
- Nakamura Y, Niki T, Goto A, et al. c-Met activation in lung adenocarcinoma tissues: an immunohistochemical analysis. Cancer Sci 2007;98:1006-13. [Crossref] [PubMed]
- Yeung SF, Tong JH, Law PP, et al. Profiling of oncogenic driver events in lung adenocarcinoma revealed MET mutation as independent prognostic factor. J Thorac Oncol 2015;10:1292-300. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Tsuta K, Kozu Y, Mimae T, et al. c-MET/phospho-MET protein expression and MET gene copy number in non-small cell lung carcinomas. J Thorac Oncol 2012;7:331-9. [Crossref] [PubMed]
- Casadevall D, Gimeno J, Clavé S, et al. MET expression and copy number heterogeneity in nonsquamous non-small cell lung cancer (nsNSCLC). Oncotarget 2015;6:16215-26. [Crossref] [PubMed]
- Schildhaus HU, Schultheis AM, Rüschoff J, et al. MET amplification status in therapy-naïve adeno- and squamous cell carcinomas of the lung. Clin Cancer Res 2015;21:907-15. [Crossref] [PubMed]
- Jurmeister P, Lenze D, Berg E, et al. Parallel screening for ALK, MET and ROS1 alterations in non-small cell lung cancer with implications for daily routine testing. Lung Cancer 2015;87:122-9. [Crossref] [PubMed]
- Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039-43. [Crossref] [PubMed]
- Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A 2007;104:20932-7. [Crossref] [PubMed]
- Gusenbauer S, Vlaicu P, Ullrich A, et al. HGF induces novel EGFR functions involved in resistance formation to tyrosine kinase inhibitors. Oncogene 2013;32:3846-56. [Crossref] [PubMed]
- Spigel DR, Ervin TJ, Ramlau RA, et al. Randomized phase II trial of Onartuzumab in combination with erlotinib in patients with advanced non-small-cell lung cancer. J Clin Oncol 2013;31:4105-14. [Crossref] [PubMed]
- Spigel DR, Edelman MJ, Mok T, et al. Treatment rationale study design for the MetLung Trial: a randomized, double-blind phase III study of onartuzumab (MetMAb) in combination with erlotinib versus erlotinib alone in patients who have received standard chemotherapy for stage IIIB or IV Met-positive non-small-cell lung cancer. Clin Lung Cancer 2012;13:500-4. [Crossref] [PubMed]
- Camidge DR, Ou SH, Shapiro G, et al. Efficacy and safety of crizotinib in patients with advanced c-MET-amplified non-small cell lung cancer (NSCLC). J Clin Oncol 2014;32:abstr 8001.
- Lennerz JK, Kwak EL, Ackerman A, et al. MET amplification identifies a small and aggressive subgroup of esophagogastric adenocarcinoma with evidence of responsiveness to crizotinib. J Clin Oncol 2011;29:4803-10. [Crossref] [PubMed]
- Kwak EL, LoRusso P, Hamid O, et al. Clinical activity of AMG 337, an oral MET kinase inhibitor, in adult patients (pts) with MET-amplified gastroesophageal junction (GEJ), gastric (G), or esophageal (E) cancer. J Clin Oncol 2015;33:abstr 1.
- Kong-Beltran M, Seshagiri S, Zha J, et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res 2006;66:283-9. [Crossref] [PubMed]
- Onozato R, Kosaka T, Kuwano H, et al. Activation of MET by gene amplification or by splice mutations deleting the juxtamembrane domain in primary resected lung cancers. J Thorac Oncol 2009;4:5-11. [Crossref] [PubMed]
- Paik PK, Drilon A, Fan PD, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov 2015;5:842-9. [Crossref] [PubMed]
- Liu X, Jia Y, Stoopler MB, et al. Next-generation sequencing of pulmonary sarcomatoid carcinoma reveals high frequency of actionable MET gene mutations. J Clin Oncol 2016;34:794-802. [Crossref] [PubMed]
- Jenkins RW, Oxnard GR, Elkin S, et al. Response to crizotinib in a patient with lung adenocarcinoma harboring a MET splice site mutation. Clin Lung Cancer 2015;16:e101-4. [Crossref] [PubMed]
- Waqar SN, Morgensztern D, Sehn J, et al. MET mutation associated with responsiveness to crizotinib. J Thorac Oncol 2015;10:e29-31. [Crossref] [PubMed]
- Mendenhall MA, Goldman JW. MET-mutated NSCLC with major response to crizotinib. J Thorac Oncol 2015;10:e33-4. [Crossref] [PubMed]


