
Integrated model for COVID-19 diagnosis based on computed tomography artificial intelligence, and clinical features: a multicenter cohort study
Introduction
Since the discovery of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) species—and the resulting novel coronavirus disease (COVID-19)—in December 2019, humanity has been plunged into a global pandemic (1). Although the devastating effects of the virus have been mitigated by vaccination (2,3), breakthrough infections caused by new variants of the virus prevent the pandemic from coming to an end (4).
The gold standard for COVID-19 diagnosis is the real-time reverse transcription polymerase chain reaction (RT-PCR) test. However, RT-PCR has several drawbacks: in several cases, it is known to be insufficiently sensitive to the virus even in symptomatic patients, leading to false negatives (5,6). In addition, its diagnosis in facilities that require specimen transport has a long turnaround time (7). These aspects reveal the need for a more accurate and timely diagnosis; for this, several diagnostic models have been developed using clinical characteristics, laboratory data, and radiographic images. However, most such models have not been validated with datasets external to the development phase (8). Moreover, methodological flaws and/or underlying biases, making it difficult to determine the model validity. Consequently, there are no diagnostic models using chest computed tomography (CT) with potential clinical use (9,10).
There is no diagnostic system that automatically interprets the CT and clinical features. In addition, to overcome the limitations of diagnostic models using chest CT, we have externally validated a deep-learning-based, CT diagnostic system for COVID-19 (Ali-M3) (11). To further improve its accuracy, it is important to properly diagnose COVID-19 patients without pneumonia detectable by CT. For this purpose, we integrated the Ali-M3 model with the clinical characteristics of patients suspected of having COVID-19 using machine learning, and validated this new system. We present the following article in accordance with the TRIPOD reporting checklist (12) (available at https://atm.amegroups.com/article/view/10.21037/atm-21-5571/rc).
Methods
We used datasets for the external validation of Ali-M3. The details of the datasets were published elsewhere (11). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The institutional review board of Hyogo Prefectural Amagasaki General Medical Center (No. 2-214) and other facility approved of our study and waived the need to obtain written informed consent.
Study design
This was a retrospective cohort study conducted in 11 Japanese tertiary care facilities that provided treatment for patients with COVID-19.
Participants
We included patients who underwent both a RT-PCR and a chest CT for the diagnosis of COVID-19. Potentially eligible participants were identified as those who had, on the advice of physicians, taken both the RT-PCR and chest CT tests when they presented with symptoms or were suspected of having COVID-19. RT-PCR results were extracted from the patients’ medical records at each facility. We selected patients by using consecutive sampling methods between January 1 and May 30, 2020. We excluded patients when the time-interval between chest CT and the first RT-PCR assay exceeded 7 days.
Chest CT and artificial intelligence (AI)
We considered, for each patient, the CT image that was taken closest to the onset of symptoms. All of these images featured the patient in a supine position.
Ali-M3 is a three-dimensional, deep-learning framework for the detection of COVID-19 infections, developed from 7,000 chest CT scans (13). It predicts COVID-19 infections with confidence levels in the range of 0–1. The learning of Ali-M3 was halted before our evaluation (11) and the investigators who entered data from the CT images into Ali-M3 were blinded to the corresponding RT-PCR results. The area under the curve (AUC) of Ali-M3 for predicting a COVID-19 diagnosis was 0.797 [95% confidence intervals (CI): 0.762 to 0.833] (11).
Clinical characteristics
We extracted, from electronic medical records, clinical characteristics that were recorded at a time closest to the date of the chest CT scans. During data acquisition, the turnaround time of RT-PCR was a few days; therefore, all predictive variables were recorded without the RT-PCR results.
Reference standard
COVID-19 was diagnosed by the RT-PCR test, which detected the presence of the nucleic acid of SARS-CoV-2 in the sputum, throat swabs, and secretions of the lower respiratory tract (14). This test was established as the primary reference standard. Although the findings of chest CT, interpreted by radiologists, were included as a reference standard in the AI development phase of this framework, we did not include it as the reference standard in this study.
Statistical analysis
Model development
We used the machine learning model, Light Gradient Boosting Machine (LightGBM), which is also a highly effective gradient-boosting decision tree algorithm (14). In the boosting algorithm, a weak classifier (decision tree) is sequentially created to minimize the prediction errors made by the previous classifier (15). The result is a powerful ensemble classifier with superior predictive performance. To avoid overfitting, parameters specific to the algorithm (known as hyperparameters) must be well tuned before fitting them to the final model, which also needs to treat missing data as such.
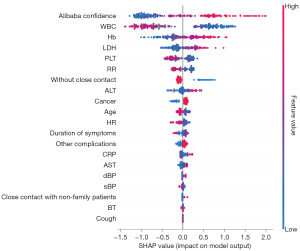
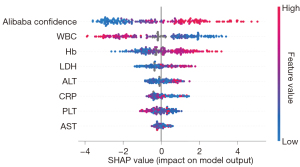
The creation of prediction models consists of three steps. First, the dataset in each hospital was chronologically split into a training set (with 70% of the patients) and a test set (with the remaining 30% of subjects). Second, hyperparameters were tuned to maximize the area under the receiver-operator curve (AUROC) by performing a fivefold cross-validation on the training set using stratified splitting in equally sized groups. A Bayesian optimization algorithm was used for tuning, with the search parameters and spaces given as follows: “num_leaves” (maximum number of leaves in one tree) at 10–150, “max_depth” (maximum tree depth) at 10–150, “learning_rate” (learning rate) at 0.005–0.5, “subsample_for_bin” (number of data sampled to construct feature-discrete bins) at 20,000–300,000, “min_child_samples” (minimal number of data in one leaf) at 10–100, “reg_alpha” (L1 regularization) at 0.0–1.0, “reg_lambda” (L2 regularization) at 0.0–1.0, “colsample_bytree” (the rate of features selected in training each tree) at 0.5–1.0, “subsample” (the rate of data selected in training each tree) at 0.5–1.0, and “is_unbalanced” with True or False. Optimal “n_estimator” (number of trees) was automatically determined by employing early stopping (“early_stopping_rounds” =100). Next, we used the entire training set to fit two final models, whose hyperparameters were also tuned. One model—the full model—included all the above-mentioned variables, while the other model—the A-blood model—included only eight limited variables (the Ali-M3 confidence variable, in addition to seven variables pertaining to blood test results: white blood cell, hemoglobin, platelet, aspartate aminotransferase, alanine aminotransferase, lactate dehydrogenase, and C-reactive protein). These blood test variables were selected owing to the ease of availability of their data and due to their relative importance in the full model, which was computed as Shapley Additive exPlanations (SHAP) values (14). SHAP values quantify the association between each variable and the outcome of each patient.
We used Python version 3.7.11 (16) with LightGBM version 2.2.3 (17) and hyperopt version 0.1.2 (18).
Model external validation
We used the temporal validation method for external validation. We differentiated between the confidence of the machine-learning models and the Ali-M3 framework by using AUROC in the test set, with 95% CIs calculated with bootstrapped resampling (1,000 samples). AUROC is an effective measure of overall diagnostic accuracy, which is deemed to be “outstanding” if AUROC ≥0.9, “excellent” if 0.8< AUROC <0.9, and “acceptable” if 0.7< AUROC <0.8 (19). Calibration was assessed using the Brier score (20) and a calibration plot. The formulation of the Brier score for a binary prediction is given by: , where the score predicts the occurrence of the outcome, ranging from 1 for an outcome that definitely occurs and 0 for one that definitely does not occur, where smaller values indicate superior model performance. The AUROC values and Brier scores of the machine-learning models were compared with those of the Ali-M3 confidence using bootstrapped resampling (1,000 samples). We calculated the SHAP values and presented them in the figures.
Results
Patient characteristics
A total of 703 patients were included in the study, including 326 PCR-positive and 377 PCR-negative patients. In the training set, we included 490 patients, including 247 PCR-positive patients. In the test set, we included 213 patients, including 79 PCR-positive patients. Patient characteristics are shown in Tables 1,2.
Table 1
| Variables | COVID-19 PCR: positive (N=326) | COVID-19 PCR: negative (N=377) |
|---|---|---|
| Age, years | 55 [43–68] | 68 [45–79] |
| Sex, male, N (%) | 197 (60.4) | 220 (58.4) |
| Smoking, current/ex-smoker, N (%) | 46 (14.1) | 64 (17.0) |
| Contact history, N (%) | ||
| With family patients | 39 (12.0) | 6 (1.6) |
| With non-family patients | 78 (23.9) | 34 (9.0) |
| None | 209 (64.1) | 337 (89.4) |
| Travel overseas, N (%) | 39 (12.0) | 14 (3.7) |
| Duration of symptom, days | 6 [4–9] | 4 [2–9] |
| Missing data, N (%) | 15 (4.6) | 15 (4.0) |
| Symptoms, N (%) | ||
| Cough | 123 (37.7) | 109 (28.9) |
| Chill | 48 (14.7) | 43 (11.4) |
| Sore throat | 81 (24.8) | 78 (20.7) |
| Diarrhea | 41 (12.6) | 25 (6.6) |
| Muscle pain | 29 (8.9) | 17 (4.5) |
| Conjunctivitis | 18 (5.5) | 12 (3.2) |
| Taste disorder | 33 (10.1) | 22 (5.8) |
| Complications, N (%) | ||
| Coronary arterial diseases | 12 (3.7) | 38 (10.1) |
| Cerebrovascular diseases | 25 (7.7) | 46 (12.2) |
| Chronic heart failures | 21 (6.4) | 59 (15.6) |
| Chronic kidney diseases | 17 (5.2) | 53 (14.1) |
| COPD | 16 (4.9) | 62 (16.4) |
| Malignancy | 29 (8.9) | 87 (23.1) |
| Immune disorders | 5 (1.5) | 28 (7.4) |
| Hypertension | 52 (16.0) | 85 (22.5) |
| Diabetes mellitus | 56 (17.1) | 84 (22.3) |
| Others | 59 (18.1) | 158 (41.9) |
| Vital signs | ||
| Body temperature, °C | 37.2 [36.6–38.1] | 37.2 [36.7–38.0] |
| Missing data, N (%) | 14 (4.3) | 25 (6.6) |
| Systolic blood pressure, mmHg | 126 [113–138] | 130 [114–148] |
| Missing data, N (%) | 20 (6.1) | 38 (10.1) |
| Diastolic blood pressure, mmHg | 79 [70–89] | 77 [67–87] |
| Missing data, N (%) | 20 (6.1) | 38 (10.1) |
| Heart rate, beats per minute | 86 [78–98] | 93 [80–108] |
| Missing data, N (%) | 11 (3.4) | 35 (9.3) |
| Respiratory rate, breaths per minute | 18 [16–21] | 20 [16–24] |
| Missing data, N (%) | 65 (19.9) | 138 (36.6) |
| Laboratory data | ||
| White blood cell, ×103/μL | 4.1 [1.8–5.2] | 9.2 [6.4–12.5] |
| Missing data, N (%) | 14 (4.3) | 44 (11.7) |
| Hemoglobin, g/dL | 14.0 [12.9–15.2] | 12.2 [10.3–13.5] |
| Missing data, N (%) | 23 (7.1) | 60 (15.9) |
| Platelet, ×104/μL | 18.9 [15.1–25.0] | 23.5 [16.4–30.0] |
| Missing data, N (%) | 16 (4.9) | 45 (11.9) |
| Aspartate aminotransferase, U/L | 32 [24–54] | 27 [19–40] |
| Missing data, N (%) | 13 (4.0) | 43 (11.4) |
| Alanine aminotransferase, U/L | 30 [17–46] | 20 [13–34] |
| Missing data, N (%) | 13 (4.0) | 43 (11.4) |
| Lactate dehydrogenase, U/L | 282 [216–403] | 244 [186–324] |
| Missing data, N (%) | 15 (4.6) | 54 (14.3) |
| C-reactive protein, mg/dL | 3.7 [0.5–9.5] | 5.5 [1.4–11.9] |
| Missing data, N (%) | 14 (4.3) | 57 (15.1) |
| Computed tomography data | ||
| Ali-M3 confidence, % | 0.93 [0.52–1.00] | 0.25 [0.01–0.71] |
All continuous variables are not normally distributed and are presented as median [interquartile range]; categorical variables are presented as N (%). COPD, chronic obstructive pulmonary disease.
Table 2
| Characteristic | H01 (N=94)1 | H02 (N=158)1 | H03 (N=19)1 | H04 (N=70)1 | H05 (N=71)1 | H06 (N=32)1 | H07 (N=21)1 | H08 (N=68)1 | H09 (N=110)1 | H10 (N=34)1 | H11 (N=43)1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age, years | 57 [44–76] | 60 [44–73] | 63 [53–76] | 60 [39–72] | 59 [44–76] | 60 [46–68] | 65 [41–76] | 63 [42–73] | 59 [42–73] | 78 [58–86] | 68 [46–80] | |
| COVID-19 PCR | ||||||||||||
| Positive | 70 (74%) | 52 (33%) | 18 (95%) | 35 (50%) | 26 (37%) | 21 (66%) | 12 (57%) | 18 (26%) | 37 (34%) | 11 (32%) | 21 (49%) | |
| System | Aquilion PRIME | Optima CT660 | Aquilion PRIME | Optima CT660 | Optima CT660 | Aquilion PRIME | Aquilion CX Edition | Aquilion ONE | Aquilion CXL | Aquilion PRIME | Aquilion CXL | Aquilion CX Edition |
| Vendor | Canon Medical Systems | GE | Canon Medical Systems | GE | GE | Canon Medical Systems | Canon Medical Systems | Canon Medical Systems | Canon Medical Systems | Canon Medical Systems | Canon Medical Systems | Canon Medical Systems |
| Tube voltage (kVp) | 120 | 120 | 120 | 120 | 120 | 120 | 120 | 120 | 120 | 120 | 120 | 120 |
| Automatic tube current modulation (mAs) | Auto | 100–510 | 150–250 | 80–500 | 80–500 | 150–250 | 403–500 | 100–400 | 100–400 | 50–250 | 100–400 | 100–400 |
| Pitch | ||||||||||||
| Standard | 111 | 55 | 65 | 55 | 55 | 65 | – | 65 | 53 | 65 | 53 | – |
| Factor | 0.813 | 0.984 | 0.813 | 0.984 | 0.984 | 0.813 | 1.172 | 0.813 | 0.828 | 0.813 | 0.828 | 1.000 |
| Matrix | 512×512 | 512×512 | 512×512 | 512×512 | 512×512 | 512×512 | 512×512 | 512×512 | 512×512 | 512×512 | 512×512 | 512×512 |
| Slice thickness (cm) | 0.500 | 0.625 | 0.500 | 0.625 | 0.625 | 0.500 | 0.500 | 0.500 | 0.500 | 0.500 | 1.000 | 5.000 |
| Field of view (mm) | 320 | 340 | 320 | – | – | 330 | 350 | 320 | 320 | 320–400 | 320 | 320 |
| Reconstruction interval (mm) | 5 | 0.625 | 2 | 1.25 | 1.25 | 3 | 5 | 5 | 5 | 5 | 5 | 5 |
1, n (%) or median [interquartile range].
Model performance
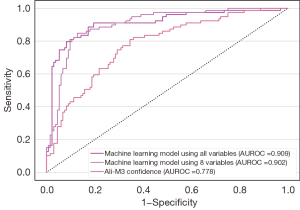
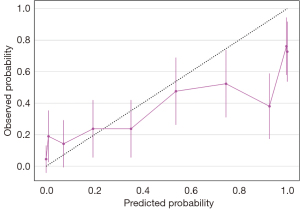
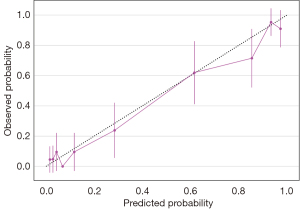
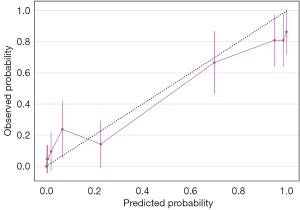
We developed two models—a full model and an A-blood model. Details of the A-blood model are accessible in web calculators online (21). We have outlined the model discrimination and calibration of the test data in Table 3. The AUROC values of both the full model (0.91, 95% CI: 0.86 to 0.95) and the A-blood model (0.90, 95% CI: 0.86 to 0.94) were better than that of the Ali-M3 confidence (0.78, 95% CI: 0.71 to 0.83) in the test set. The calibration evaluated by the Brier scores of both the full model (0.10, 95% CI: 0.07 to 0.13) and the A-blood model (0.12, 95% CI: 0.08 to 0.16) were better than that of the Ali-M3 confidence (0.23, 95% CI: 0.19 to 0.27) in the test set. The ROCs of the test data are shown in Figure 1, with SHAP values shown in Figures 2,3. Figure 2 shows all of the predictive variables that were used. Figures 4-6 show the calibration plots for the Ali-M3 framework, the full model, and the A-blood model, respectively.
Table 3
| Model | Score | Difference between Ali-M3 confidence | |||
|---|---|---|---|---|---|
| AUROC (95% CI) | Brier score (95% CI) | AUROC (95% CI) | Brier score (95% CI) | ||
| Full model* | 0.91 (0.86 to 0.95) | 0.10 (0.07 to 0.13) | 0.13 (0.07 to 0.19) | −0.13 (−0.17 to −0.09) | |
| A-blood model | 0.90 (0.86 to 0.94) | 0.12 (0.08 to 0.16) | 0.12 (0.07 to 0.18) | −0.11 (−0.15 to −0.07) | |
| Ali-M3 confidence | 0.78 (0.71 to 0.83) | 0.23 (0.19 to 0.27) | – | − | |
*, machine learning model using all variables; , machine learning model using 8 variables including Ali-M3 confidence, white blood cell, hemoglobin, platelet, aspartate aminotransferase, alanine aminotransferase, lactate dehydrogenase, and C-reactive protein. AUROC, area under receiver operator curve; CI, confidence interval.
Discussion
We have developed and validated two integrated diagnostic models of the Ali-M3 framework with the clinical characteristics of patients with suspected COVID-19. Based on the relative importance of each variable, we shrank the full model to a more compact A-blood model, whose parameters included the Ali-M3 confidence and eight routinely collected blood markers. This A-blood model showed a better discrimination and calibration performance than the full model.
Our diagnostic model is the first to automatically interpret clinical data in conjunction with CT scans. Several problems faced by existing diagnostic models, such as the separate collection of cases and controls, the lack of external validation, and insufficient reporting (8,9), have been overcome in this study with rigorous methodology, with our model achieving good discrimination and calibration performance.
The A-blood model may allow for quicker diagnoses at emergency departments. Even if the RT-PCR test is available in the facility, the A-blood model might be a better option because of its lower turnaround time, which requires only a general blood test and CT results. In the majority of Japanese emergency hospitals, including the 11 hospitals in the dataset, the time to CT imaging for stroke patients is less than 20 min (22). Even during the COVID-19 pandemic, no substantial increase was observed in the time to obtain CT (23). When the RT-PCR results are not known, the “A-blood” model could help physicians determine indications for timely treatment with antibody drugs (24). For patients for whom COVID-19 infection cannot be ruled out based on a single negative RT-PCR, physicians may be able to use the “A-blood” model to determine if a patient can be released from quarantine. These clinical implications need to be evaluated in further studies (25).
This study has several limitations. First, the dataset used in this study is from the first wave of infections in the spring of 2020, which does not include vaccinated patients or the latter variants of the SARS-CoV2 virus. Therefore, it is necessary to further expand on this external validation. A second limitation is the occurrence of false negatives, which includes patients falsely regarded as COVID-negative with a single negative PCR result. This misclassification may affect the accuracy, but the magnitude of this bias cannot be predicted. Further studies are required with datasets that also include sufficient follow-up. Third, because of the retrospective nature, we could not define rigorous inclusion criteria, such as symptoms or settings.
In conclusion, we developed the A-blood model, which is a COVID-19 diagnostic tool that combines machine learning and CT evaluation with blood test data. Physicians would be able to use this model for the rapid diagnosis of COVID-19. Further validation studies, especially those including SARS-CoV-2 variants and subjects inoculated with different vaccines, are warranted.
Acknowledgments
We thank M3 Inc. and Clinical Porter for providing free Ali-M3 analysis and data storage, although they did not participate in the preparation of the protocol and the manuscript. To access Ali-M3, the reader can contact M3 (m3-ai-lab@m3.com). We thank Editage (www.editage.jp) for the English language review. We also thank Ms. Kyoko Wasai, who assisted in retrieving the data.
Funding: This study was partially supported by Kyoto University managing fund for English editing. The article processing fee was supported by the Scientific Research Works Peer Support Group (SRWS-PSG). Funders played no role in the design or conduct of the study; the collection, management, analysis, or interpretation of the data; the preparation, review, or approval of the manuscript; or the decision to submit the manuscript for publication.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-5571/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-21-5571/dss
Peer Review File: Available at https://atm.amegroups.com/article/view/10.21037/atm-21-5571/prf
Conflicts of Interest: All authors have completed the ICMJE unified disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-5571/coif). Yuki Kataoka serves as an unpaid editorial board member of Annals of Translational Medicine from July 2020 to June 2022. Yuki Kataoka, Yuya Kimura, TI, YM, JM, JK, KT, HF, TH, AK, FH, SI, SF report that this study was partially supported by Kyoto University managing fund for English editing; the article processing fee was supported by the Scientific Research Works Peer Support Group (SRWS-PSG); M3 Inc. and Clinical Porter provided free Ali-M3 analysis and data storage. TI also reports that the analysis of the CT by Ali-M3 was carried out by Nobori on behalf of M3. M3 and Nobori did not know the patients’ data including the result of RT-PCR. JM also reports that receiving a lecture fee from M3 Inc. The other author has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The institutional review board of Hyogo Prefectural Amagasaki General Medical Center (No. 2-214) and other facility approved of our study and waived the need to obtain written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Carvalho T, Krammer F, Iwasaki A. The first 12 months of COVID-19: a timeline of immunological insights. Nat Rev Immunol 2021;21:245-56. [Crossref] [PubMed]
- Baden LR, El Sahly HM, Essink B, et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med 2021;384:403-16. [Crossref] [PubMed]
- Polack FP, Thomas SJ, Kitchin N, et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med 2020;383:2603-15. [Crossref] [PubMed]
- Bergwerk M, Gonen T, Lustig Y, et al. Covid-19 Breakthrough Infections in Vaccinated Health Care Workers. N Engl J Med 2021;385:1474-84. [Crossref] [PubMed]
- Watson J, Whiting PF, Brush JE. Interpreting a covid-19 test result. BMJ 2020;369:m1808. [Crossref] [PubMed]
- Kortela E, Kirjavainen V, Ahava MJ, et al. Real-life clinical sensitivity of SARS-CoV-2 RT-PCR test in symptomatic patients. PLoS One 2021;16:e0251661. [Crossref] [PubMed]
- Kohli A, Joshi A, Shah A, et al. Does CT help in reducing RT-PCR false negative rate for COVID-19? Indian J Radiol Imaging 2021;31:S80-6. [Crossref] [PubMed]
- Wynants L, Van Calster B, Collins GS, et al. Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal BMJ 2020;369:m1328. [Crossref] [PubMed]
- Roberts M, Driggs D, Thorpe M, et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat Mach Intell 2021;3:199-217. [Crossref]
- Alsharif MH, Alsharif YH, Yahya K, et al. Deep learning applications to combat the dissemination of COVID-19 disease: a review. Eur Rev Med Pharmacol Sci 2020;24:11455-60. [PubMed]
- Ikenoue T, Kataoka Y, Matsuoka Y, et al. Accuracy of deep learning-based computed tomography diagnostic system for COVID-19: A consecutive sampling external validation cohort study. PLoS One 2021;16:e0258760. [Crossref] [PubMed]
- Collins GS, Reitsma JB, Altman DG, et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): the TRIPOD statement. Ann Intern Med 2015;162:55-63. [Crossref] [PubMed]
- Academy TAD. Assisted Analysis Based on Chest CT Imaging. Alibaba DAMO Academy 2020;2.
- Kucirka LM, Lauer SA, Laeyendecker O, et al. Variation in False-Negative Rate of Reverse Transcriptase Polymerase Chain Reaction-Based SARS-CoV-2 Tests by Time Since Exposure. Ann Intern Med 2020;173:262-7. [Crossref] [PubMed]
- Friedman JH, Popescu BE. Importance Sampled Learning Ensembles. 2003.
- Welcome to Python.org [cited 2021 Sep 10]. Available online: https://www.python.org/
- GitHub. microsoft/LightGBM: A fast, distributed, high performance gradient boosting (GBT, GBDT, GBRT, GBM or MART) framework based on decision tree algorithms, used for ranking, classification and many other machine learning tasks. [cited 2021 Sep 10]. Available online: https://github.com/Microsoft/LightGBM
- Hyperopt Documentation [cited 2021 Sep 10]. Available online: http://hyperopt.github.io/hyperopt/
- Mandrekar JN. Receiver operating characteristic curve in diagnostic test assessment. J Thorac Oncol 2010;5:1315-6. [Crossref] [PubMed]
- Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology 2010;21:128-38. [Crossref] [PubMed]
- A-blood model web calculator [cited 2021 Oct 14]. Available online: https://youkiti.shinyapps.io/ablood-for-Dx-COVID-19/
- Okada T, Inoue M, Yamagami H, et al. Nationwide questionnaire survey on neuroimaging strategy for acute ischemic stroke in Japan. Japanese Journal of Stroke 2020;42:502-8. [Crossref]
- Koge J, Shiozawa M, Toyoda K. Acute Stroke Care in the With-COVID-19 Era: Experience at a Comprehensive Stroke Center in Japan. Front Neurol 2021;11:611504. [Crossref] [PubMed]
- Siemieniuk RA, Bartoszko JJ, Díaz Martinez JP, et al. Antibody and cellular therapies for treatment of covid-19: a living systematic review and network meta-analysis. BMJ 2021;374: [Crossref] [PubMed]
- Lijmer JG, Leeflang M, Bossuyt PM. Proposals for a phased evaluation of medical tests. Med Decis Making 2009;29:E13-21. [Crossref] [PubMed]


