MicroRNAs in congenital heart disease
Introduction
The discovery of non-coding RNAs has provided new insight into the mechanisms that underpin human congenital and acquired diseases. This review will focus in microRNAs (miRs) and congenital heart disease (CHD).
miRs are small, evolutionally conserved, non-coding RNA molecules which have been shown to negatively regulate gene expression (1). Initially identified in animals they are now recognised to be widely distributed in the eukaryotic kingdom and are commonly found in vertebrates. It is estimated that in excess of 1,000 miRs are expressed in humans Furthermore, bioinformatic analyses suggests that the miRs have the potential to regulate 30% of human genes through a series of complex signalling pathways (2). Moreover, miRs can co-ordinately regulate the stability of multiple target genes. Thus, aberrant expression of miRs can affect multiple intracellular signalling pathways and are associated with many diseases such as cancer, diabetes and heart disease (3-5).
Furthermore, miRs are now known to be key components to the embryonic development of the heart, normal cardiovascular function and cardiac pathophysiology in multiple cell lineages (6-12).
The Global Burden of Disease Study [2013] estimated that almost 30% of all deaths worldwide were caused by cardiovascular disease (13). Along the spectrum of adult cardiovascular diseases, congenital pathology is often the aetiology. Therefore, a new approach to the identification and treatment of CHD is necessary to reduce the prevalence of disease in young and old populations. Of all congenital malformations, CHD comprises of the majority of cases with a prevalence rate of 8 in every 1,000 infants (14). Over 40% of prenatal deaths can be attributed to CHD (15). The incidence of CHD has been associated with both increased neonatal and maternal morbidity. The prevalence of CHD varies widely and is more diffuse in Europe than Northern America (16,17). The Euro Heart Survey suggests that up to 19% of patients with CHD undergo surgery or a catheter-based intervention (18). Common CHD phenotypes include, atrial septal defect (ASD), ventricular septal defect (VSD), patent ductus arteriosus (PDA), tetralogy of Fallot (TOF), transposition of the great arteries (TGA), pulmonary valve atresia (PA), coarctation of the aorta (COA) and tricuspid atresia (TA) (see Table 1 for a summary of CHD types). Furthermore, these phenotypes can also manifest in syndromic patterns with genetic associations, such as atrioventricular septal defects (AVSD) in patients with Down Syndrome (20).
This review will evaluate the evidence for the association of miRs in CHD, explore the cause-effect relationship in disease states and discuss their potential as therapeutic biomarkers.
miR biogenesis: an overview
The following paragraph will be an overview of miR biogenesis, for more detail we recommended a reading by Gama-Carvalho et al. (21), miR biogenesis begins with a long 5’-capped and poly A tailed, primitive form of miRNA (pri-miR) transcript configured into a hairpin structure which are derived from protein coding genes or independent non-coding transcriptional unit (22,23). These miR producing genes or are transcriptionally regulated like other protein coding genes but often contain polycistronic clusters.
Maturation of pri-miRs is initiated in the nucleus of a cell to produce precursor miR (pre-miR) which is transported to the cytosol or the endoplasmic reticulum to be cut into its mature form (approximately 22 nucleotides long) by Dicer, a RNase III endonuclease. Dicer activity is critical to miRNA biogenesis and impacts cardiac physiology. In an attempt to investigate the biological importance of miRNAs, mutation or disruption of Dicer has been employed by various groups as a broad method to prevent miRNAs production. Both in vitro and in vivo, evidence exists to support a role for Dicer-dependent miRNAs in vascular signaling and multi-system roles related to angiogenesis (24-28). Indeed, selective deletion of Dicer impacts the regulation of cardiac morphogenesis, electrical conduction, and cell-cycle control (8). In addition, dilated cardiomyopathy associated with heart failure, and spontaneous cardiac remodeling is found with the deletion of Dicer (29,30). More broadly, Suárez et al. excellently review the literature on miRs in the regulation of angiogenesis, with specific mention to dicer selective knockout models in cellular and animal models (31).
The mature miR is a single strand of RNA which has the potential to be recruited to the RNA-induced silencing complex (RISC), which also comprises Argonaute (Ago) proteins. In the RISC, the miR can repress the expression of its messenger RNA (mRNA) targets. Each miR has the potential to repress the expression of multiple genes. miR achieves this by first recognising a complimentary (or semi complimentary) “seed sequence” containing 8 nucleotides in the 5' untranslated region (5'UTR) to miR binding sites of the 3' untranslated region (3'UTR) of the target mRNA. Ultimately, the targeted mRNA repression can be achieved by mRNA degradation, transcript deadenylation, translation inhibition or sequestration of the mRNAs in the processing body (P-body) (32). miRs can also be released extracellularly and are present in virtually any biological fluid. In comparisons to mRNAs, miRs are more resistant to degradation because of several mechanisms of protection, for example their being engulfed within extracellular vesicles or conjugated to lipoproteins or Ago proteins (33). Table 2 summarized the miRs so far implicated in development of CHD.
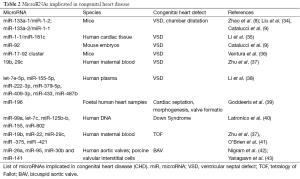
Full table
Ventricular septal defects (VSD)
A VSD is a discontinuation in the septal wall dividing the left and right ventricles of the heart. VSDs may be present at birth or can be acquired after myocardial infarction. VSDs account for approximately 20–40% of CHD but 80% of the surgical workload (33,44). Large defects may present with sever heart failure in infancy. However, small defects may remain asymptomatic. VSDs lead to a left to right shut of circulation producing left ventricular volume overload resulting in pulmonary hypertension (45).
MiR-1-1 and miR-181c have been implicated in the pathogenesis of VSDs (35). MiR-1 is a regulator of bone morphogenic protein receptor type II (BMPR2) and gap junction protein alpha 1 (GJA1) while miR-181c can regulate sex determining region Y (SRY)-box 9 (SOX9). In human cardiac tissue with VSDs, elevated levels of GJA1 and SOX9 coincided with reduced expression of miR-1-1, and elevated miR-181c expression was associated with down regulation of BMPR2 (35).
Over-expression of miR-1 plays a fundamental role in ventricular cardiomyocyte proliferation and prevents expansion of the ventricular myocardium (46). Hand2 (a transcription factor that promotes ventricular cardiomyocyte expansion) is a target for miR-1. In addition, this study showed that knockouts of miR-1 results in a reduced pool of proliferating ventricular cardiomyocytes mass in the developing heart (46). Furthermore, haplo insufficiency of miR-1 or miR-133a is associated with an increased risk of VSD via a process of Hand2 and serum response factor (SRF) respectively (9). In addition to this, a reduction in miRNA-1 and miR-133 expression is associated with cardiac hypertrophy in murine models and human diseases associated with cardiac hypertrophy (9). Similarly, miR-92 deficiency is associated with VSDs in mouse embryos (9).
Targeted deletion of the miR-1-2 gene in mice produces 50% embryonic lethality as a result of VSDs. The surviving miR-1-2 homozygous mice exhibited a diverse range of phenotypes including, rapid dilation of the heart and ventricular dysfunction with of atrial thrombi (8). MiR-133a-1/miR-1-2 and miR-133a-2/miR-1-1 are myocyte enhancer factor (MEF)-2 dependant enhancers which have been shown to be activate in the linear heart tube during mouse embryogenesis and controls transcription in the cardiac chambers (34). Both miR-133a-1/miR-1-2 and miR-133a-2/miR-1-1 genes are expressed in the intraventricular septum and the ventricular myocardium (9,34). Interestingly, singular deletion of either miR-133a-2 or miR-133a-1 in mice does not result in pathology. However, combined deletion of miR-133a-2 and miR-133a-1 produced late embryonic and neonatal deaths due to VDS and dilatation of the cardiac chambers (47). Similarly, targeted deletion of the miR-17-92 family of miRs results in neonatal lethality from lung hypoplasia and VSDs (36).
A Chinese study, investigated circulating miR in 20 patients with VSDs compared with 8 VSD-free controls (38). This group identified 1 miR significantly up-regulated (hsa-miR-498) and 7 miRs which were down-regulated in the VSD group (let-7e-5p, miR-155-5p, miR-222-3p, miR-379-5p, miR-409-3p, miR-433, miR-487b). Gene ontology analysis in this study suggested that right ventricle morphogenesis were the potential target of these miRs. Specifically, this group predicted NOTCH1, HAND1, ZFPM2, and GATA3 as mRNA targets of let-7e-5p, miR-222-3p and miR-433 (39).
Foetal human heart samples have been found to contain mir-196a at gestational age 12–14 weeks (48). Mir-196a is implicated in HOXB8-Shh signalling which is utilised throughout cardiac septation, morphogenesis and valve formation (48). Therefore, miR-196a dysregulation could have a role in the formation of atrioventricular septal defects (AVSDs) and cardiac valve dysfunction.
Syndromic congenital heart disease (CHD)
In a population not affected by prenatal diagnosis, 40–60% of babies born with Down syndrome have CHD (49,50). Downs syndrome is characterised by a number of clinical signs and symptoms, it is often diagnosed via fluorescence in situ hybridisation (FISH) which demonstrates trisomy of chromosome 21. Downs syndrome has now been linked to five miRs, including, miR-99a, let-7c, miR-125b-2, miR-155 and miR-802 (40). These miRs have been identified on human chromosome 21. In addition, these miRs have been found to be over-expressed in cardiac tissue of patients with trisomy 21 (40). Furthermore, DiGeorge syndrome which results from the deletion of critical region 8 on chromosome 22 (22q11.2) is responsible for the encoding a component of the RNA-induced slicing complex essential for miR biogenesis, leading to haploinsufficiency of this complex (51). Many patients with DiGeorge Syndrome have associated CHD. This association suggests that multiple miRs are implicated in this syndrome and that dysfunction of miRNA expression could contribute to a gene dosage sensitivity to this disease (51,52).
Embryological links between cardiac and neuronal-craniofacial defects exits at the molecular level and clinically. Deletions of Dicer in neural crest cells result in a range of sever cardio-facial-crest defects. These syndromes include Noonans Syndrome, DiGeorge Syndrome, LEOPARD syndrome, cardio-facio-cutaneous syndrome and Costello syndrome (53-55).
Cyanotic congenital heart disease (CHD)
More recently, miRs have been investigated into the aetiology of cyanotic CHD (41). TOF is the most common form of cyanotic CHD and represents 5–7% of all CHD, with males and females equally affected (56-58). The term TOF describes the tetrad of (I) mostly large and non-restrictive VSDs; (II) an over-riding aorta; (III) right ventricular outflow obstruction; and (IV) right ventricular hypertrophy (59). TOF is now recognised as a spectrum of diseases which share similar intracardiac pathology. The exact cause of TOF is unknown. However, there is a growing understanding of the importance of 22q11 in its incidence. For example, Di George syndrome and velocardiofacial syndrome, both of which have 22q11 deletions, is frequently co-diagnosed in those with TOF (60).
O’Brien et al. identified an association with non-syndromic TOF, miRs and spliceosomal RNAs (41). This group identified 61 miRs to have significant changes in expression levels in children with TOF compared with normally developing children. Interestingly, the levels of miR expression in children with TOF remained similar to those in the foetal myocardium. This group looked at gene expression critical to cardiac development and their correlation to miR expression in TOF myocardium. They found that in children with TOF, splicing variants were observed in 51% of genes critical to cardiac development. They identified 33 miRs which were significantly down-regulated in TOF myocardial tissue compared to the normally developing myocardium (41). Together these findings suggest central roles for miRNAs and their spliceosomal function in TOF.
Later this group identified an inverse correlation between the expression of miR-421 and SOX4 in patients with TOF. SOX4 is a key regulator of the Notch pathway, which has been implicated in cardiac function, suggesting that miR-421 is a potential contributor to TOF (61).
Bicuspid aortic valve (BAV)
BAV are a leading cause of calcific aortic stenosis and insufficiency which results in a high prevalence of thoracic aortic aneurysms in this patient group, BAV is a common congenital cardiac defect which has a population presence of 1–2% (62).
Recently, Yanagawa et al. have identified distinct miR profiles a small cohort of human BAV leaflets in comparisons with control patients with a tricuspid aortic valve (TAV). This group identified 8 miRs which were up-regulated and 27 miRs which were down-regulated in patients with BAV, compared to patients with TAV. Most significantly, expression of miR-141 was down-regulated 14.5 fold in patients with BAV (43).
Nigam et al. further investigated the association of miRs and BAV (42). In this study, the authors investigated miR expression in aortic valve leaflets of patients with aortic stenosis and those with aortic insufficiency in nine patients undergoing aortic valve replacement. They were able to show that miR-26a and miR-195 levels were significantly reduced and miR-30b expression to be reduced by 62% (P<0.06) using quantitative reverse transcription-polymerase chain reaction. Following this they identified that human aortic valve interstitial cells treated with miR-26a or miR-30b mimics reduced miR levels of calcification-related genes, such as BMP2, alkaline phosphatase (ALPL) and SMAD1 and of SMAD3. Interestingly, aortic valve interstitial cells treated with miR-195 showed increased mRNA levels of calcification-related genes, specifically BMP2 and RUNX2.
miR-mediated regulation in CHD
miR mediated signalling in the formation of CHD may include multiple pathways. Intracellular signalling activated by transforming growth factor beta (TGF-β) have a key role in cardiovascular development and specifically in cardiogenesis. Studies in both humans and animal models have indicated that altered TGF-β activity results in a variety of CHDs including, double outlet right ventricle, septal defects and an overriding tricuspid valve (63,64). Although not essential for cardiac development, inactivation of the genes encoding the TGF-β type 1 (TGFBR1) or type 2 receptors (TGFBR2) in cardiac myocytes leads to severe valvuloseptal defects (65,66). Interestingly, inactivation in cardiomyocytes was not shown to lead to obvious cardiac defects in embryos by this group. Transgenic evidence suggests that constitutively activated TGFBR1 arrests cardiac development at the looping stage and results in ventricular hypoplasia (67). In addition, human genetic studies have supported the significance of altered TGF-β signalling in CHD. For example, mutations in the genes encoding for TGFBR1 and TGFBR2 are associated with Marfan syndrome and Loeys-Dietz syndrome, both of which are implicated in CHD (68-71). Furthermore, there is evidence to show that mutations in TGFB2 and SMAD3 are associated with syndromic aortic aneurysms (72-74).
Peng et al. have shown that inactivated Dicer1 in mice at midgestation leads to severe myocardial wall defects (75). These mutant hearts display abnormal cell proliferation, apoptosis, and expression of contractile proteins. Expression of TGFBR1 is up-regulated in mutant hearts and inhibition of TGFBR1 reduces the defect observed in cardiomyocyte apoptosis. To add another layer of complexity, TGFBR1 mRNA is regulated by multiple miRNAs at different stages of cardiogenesis (75-77).
In human cardiac tissue, Akt is highly expressed. Akt is a protein which is known to have critical application in the regulation of cardiac development including proliferation, metabolism, angiogenesis and survival through a process of phosphorylation of downstream substrates that control the apoptotic machinery (78-80). Akt mediated signalling is complex, and involves a system of miRs, PIWIs (P-element-induced wimpy testis) interacting RNAs (piRNAs) and their associated proteins (78-83). A In the embryonic heart Akt3 is highly expressed, whereas Akt1 is predominantly expressed in the adult heart (79).
As previously discussed, abnormal miR-155 activity is implicated in patients with Down Syndrome. A study investigating miR-155 in human cardiomyocyte progenitor cells has showed that increased expression of miR-155 can inhibit necrosis. However, they observed that necrotic cell death was not induced by inhibiting endogenous miR-155. Their study also suggested that increased miR-155 levels did not impact the expression patterns of cell survival and apoptotic related genes. Therefore, miR-155 inhibits necrosis mediated by repressing the receptor interacting protein 1 (RIP1), but independently of the Akt pro-survival pathway (81).
Other miRs known to be implicated in CHD have also been linked to the Akt signalling pathway. For example, miR-92 is thought to activate the Akt pathway through inhibiting its negative regulator PHLPP2 (84). MiR-92 increases resistance to apoptosis and deficiency of miR-92 resulting in apoptosis, which may induce the formation of the VSD phenotype (84). The miR-17-92 cluster which is highly expressed in the murine myocardium may protect the heart by diminishing the apoptosis and alleviating ischemia (84). Furthermore, overexpression of MiR-1 targets Akt, via an insulin sensitive pathway which may be partially responsible for the formation of VSDs (85,86). MiR-26a and miR-22 targets PTEN leading to activation of Akt which may precipitate complex CHD, including TOF and BAV (87-89).
MiR mediated signalling is likely to be complex and driven by multiple factors (79). However, this evidence suggests that miR mediated signalling in the myocardium may provide critical information leading to novel therapeutic targets in CHD.
miRs as a biomarker
MiRs are attractive clinical biomarkers as they remain stable in blood, urine and other biological fluids and evade RNA degrading enzymes (90-93). After using sequencing by oligonucleotide ligation and detection (SOLiD) sequencing to systemically screen maternal serum miRNAs, Zhu et al. hypothesised that miRs in the maternal serum could act as a candidate biomarker for the prenatal detection of foetal CHD in early pregnancy (37). This group studied 60 women in total (30 control women with normal pregnancies and 30 pregnant women who have foetuses with CHD) and identified four significantly up-regulated miRs (miR-19b, miR-22, miR-29c, miR-375) in mothers carrying foetuses with CHD. Sensitivity for these biomarkers ranged from 55.6–77.8% and specificity ranged from 66.7–88.9%. Furthermore, a combination of the four differentially expressed biomarkers was showed to be a more efficient marker for CHD detection. Of note, miR-19b and miR-29c were significantly up-regulated in VSDs and all four miRs were up-regulated in TOF. Furthermore, miR-22 may be specifically upregulated in TOF. The results of this study are very important because they suggest that specific miR are associated with types of CHD, furthermore they explore the use of serum detection is a possible method for prenatal diagnosis. However, this idea is its infancy and there are certainly some limitations to this study regarding the sample size, huge heterogeneity of CHD and possibly variability within the mother populations themselves. Further research is required to accurately explore the possibility that miR can be used in the clinical practice for prenatal detection in CHD.
Discussion
The aetiology of CHD is likely to be a multifactorial process with contributions from anomalous gene expression and processing, epigenetic factors and a variety of environmental factors. It is considered that miRs over and under expression and co-expression have specific and generalised effects on cell signalling pathways involved in CHD. Despite our expanding knowledge base of the genetic basis and signalling pathways involved in vertebrate cardiac formation there are still huge gaps that require further investigation.
Previous studies have identified a central role for miRs in embryonic cardiogenesis (e.g., miR-1 and mir-133-a/b). However, it is likely that miRs have multiple effects in embryology across different cell linages and also in disease progression.
In light of recent advances in our knowledge base regarding miR expression and function in human and animal studies, there are still significant roles of miRs in physiology and pathophysiological process we have yet to discover.
It is hoped that a simple blood or urine test may be a novel diagnostic biomarker for the detection of CHD. miR detection from placental tissues from foetuses with CHD and from maternal peripheral blood suggests a role for serum biomarkers as an early way to detect such CHD. Measuring these abundant molecules in minimally-invasive tests on easily accessible maternal and children samples may provide highly specific and sensitive future role in the prenatal and postnatal detection of CHD.
Acknowledgements
This work was funded by the British Heart Foundation (BHF) Programme grant “MicroRNAs from cardiac surgery to basic science—and back?” (to CE), the BHF Chair in Cardiovascular Science Research programme (to CE), the Leducq transatlantic network in vascular microRNAs (MIRVAD) (to CE) and the National Institute of Health Research (NIHR) Bristol Cardiovascular Biomedical Research Unit (BRU) (to MC and CE). TS was awarded a 2014 INSPIRE summer studentship by the University of Bristol. The views expressed are those of the Authors and not necessarily those of the NHS, the NIHR or the Department of Health.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [PubMed]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15-20. [PubMed]
- Trionfini P, Benigni A, Remuzzi G. MicroRNAs in kidney physiology and disease. Nat Rev Nephrol 2015;11:23-33. [PubMed]
- Simpson LJ, Ansel KM. MicroRNA regulation of lymphocyte tolerance and autoimmunity. J Clin Invest 2015;125:2242-9. [PubMed]
- Chen J. Signaling pathways in HPV-associated cancers and therapeutic implications. Rev Med Virol 2015;25:24-53. [PubMed]
- Cai B, Pan Z, Lu Y. The roles of microRNAs in heart diseases: a novel important regulator. Curr Med Chem 2010;17:407-11. [PubMed]
- Heidenreich PA, Trogdon JG, Khavjou OA, et al. Forecasting the future of cardiovascular disease in the United States: a policy statement from the American Heart Association. Circulation 2011;123:933-44. [PubMed]
- Zhao Y, Ransom JF, Li A, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 2007;129:303-17. [PubMed]
- Catalucci D, Latronico MV, Condorelli G. MicroRNAs control gene expression: importance for cardiac development and pathophysiology. Ann N Y Acad Sci 2008;1123:20-9. [PubMed]
- Thum T, Catalucci D, Bauersachs J. MicroRNAs: novel regulators in cardiac development and disease. Cardiovasc Res 2008;79:562-70. [PubMed]
- Chen J, Wang DZ. microRNAs in cardiovascular development. J Mol Cell Cardiol 2012;52:949-57. [PubMed]
- Yu ZB, Han SP, Bai YF, et al. MicroRNA expression profiling in fetal single ventricle malformation identified by deep sequencing. Int J Mol Med 2012;29:53-60. [PubMed]
- GBD 2013 Mortality and Causes of Death Collaborators. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015;385:117-71. [PubMed]
- Meberg A, Lindberg H, Thaulow E. Congenital heart defects: the patients who die. Acta Paediatr 2005;94:1060-5. [PubMed]
- Trojnarska O, Grajek S, Katarzyński S, et al. Predictors of mortality in adult patients with congenital heart disease. Cardiol J 2009;16:341-7. [PubMed]
- Campbell M. Incidence of cardiac malformations at birth and later, and neonatal mortality. Br Heart J 1973;35:189-200. [PubMed]
- van der Linde D, Konings EE, Slager MA, et al. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol 2011;58:2241-7. [PubMed]
- Engelfriet P, Boersma E, Oechslin E, et al. The spectrum of adult congenital heart disease in Europe: morbidity and mortality in a 5 year follow-up period. The Euro Heart Survey on adult congenital heart disease. Eur Heart J 2005;26:2325-33. [PubMed]
- EUROCAT Special Report on Congenital Heart Defects. EUROCAT Central Registry, University of Ulster. Available online: . Accessed November 03, 2015.http://www.eurocat-network.eu/content/Special-Report-CHD.pdf
- Tubman TR, Shields MD, Craig BG, et al. Congenital heart disease in Down's syndrome: two year prospective early screening study. BMJ 1991;302:1425-7. [PubMed]
- Gama-Carvalho M, Andrade J, Brás-Rosário L. Regulation of Cardiac Cell Fate by microRNAs: Implications for Heart Regeneration. Cells 2014;3:996-1026. [PubMed]
- Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004;10:1957-66. [PubMed]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, et al. Identification of mammalian microRNA host genes and transcription units. Genome Res 2004;14:1902-10. [PubMed]
- Kuehbacher A, Urbich C, Zeiher AM, et al. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res 2007;101:59-68. [PubMed]
- Suárez Y, Fernández-Hernando C, Pober JS, et al. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res 2007;100:1164-73. [PubMed]
- Suárez Y, Fernández-Hernando C, Yu J, et al. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci U S A 2008;105:14082-7. [PubMed]
- Yang WJ, Yang DD, Na S, et al. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem 2005;280:9330-5. [PubMed]
- Shilo S, Roy S, Khanna S, Sen CK. Evidence for the involvement of miRNA in redox regulated angiogenic response of human microvascular endothelial cells. Arterioscler Thromb Vasc Biol 2008;28:471-7. [PubMed]
- Chen JF, Murchison EP, Tang R, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A 2008;105:2111-6. [PubMed]
- da Costa Martins PA, Bourajjaj M, Gladka M, et al. Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation 2008;118:1567-76. [PubMed]
- Suárez Y, Sessa WC. MicroRNAs as novel regulators of angiogenesis. Circ Res 2009;104:442-54. [PubMed]
- Emanueli C, Shearn AI, Angelini GD, et al. Exosomes and exosomal miRNAs in cardiovascular protection and repair. Vascul Pharmacol 2015;71:24-30. [PubMed]
- Hoffman JI. Incidence of congenital heart disease: I. Postnatal incidence. Pediatr Cardiol 1995;16:103-13. [PubMed]
- Liu N, Williams AH, Kim Y, et al. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci U S A 2007;104:20844-9. [PubMed]
- Li J, Cao Y, Ma XJ, et al. Roles of miR-1-1 and miR-181c in ventricular septal defects. Int J Cardiol 2013;168:1441-6. [PubMed]
- Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 2008;132:875-86. [PubMed]
- Zhu S, Cao L, Zhu J, et al. Identification of maternal serum microRNAs as novel non-invasive biomarkers for prenatal detection of fetal congenital heart defects. Clin Chim Acta 2013;424:66-72. [PubMed]
- Li D, Ji L, Liu L, et al. Characterization of circulating microRNA expression in patients with a ventricular septal defect. PLoS One 2014;9:e106318. [PubMed]
- Goddeeris MM, Rho S, Petiet A, et al. Intracardiac septation requires hedgehog-dependent cellular contributions from outside the heart. Development 2008;135:1887-95. [PubMed]
- Latronico MV, Catalucci D, Condorelli G. MicroRNA and cardiac pathologies. Physiol Genomics 2008;34:239-42. [PubMed]
- O'Brien JE Jr, Kibiryeva N, Zhou XG, et al. Noncoding RNA expression in myocardium from infants with tetralogy of Fallot. Circ Cardiovasc Genet 2012;5:279-86. [PubMed]
- Nigam V, Sievers HH, Jensen BC, et al. Altered Micrornas in Bicuspid Aortic Valve: A Comparison between Stenotic and Insufficient Valves. J Heart Valve Dis 2010;19:459-65. [PubMed]
- Yanagawa B, Lovren F, Pan Y, et al. miRNA-141 is a novel regulator of BMP-2-mediated calcification in aortic stenosis. J Thorac Cardiovasc Surg 2012;144:256-62. [PubMed]
- Waight DJ, Bacha EA, Kahana M, et al. Catheter therapy of Swiss cheese ventricular septal defects using the Amplatzer muscular VSD occluder. Catheter Cardiovasc Interv 2002;55:355-61. [PubMed]
- Anderson RH, Brown NA, Mohun TJ. Insights regarding the normal and abnormal formation of the atrial and ventricular septal structures. Clin Anat 2015. [Epub ahead of print]. [PubMed]
- Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 2005;436:214-20. [PubMed]
- Liu N, Bezprozvannaya S, Williams AH, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev 2008;22:3242-54. [PubMed]
- Thum T, Galuppo P, Wolf C, et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation 2007;116:258-67. [PubMed]
- Torfs CP, Christianson RE. Anomalies in Down syndrome individuals in a large population-based registry. Am J Med Genet 1998;77:431-8. [PubMed]
- Frid C, Drott P, Lundell B, et al. Mortality in Down's syndrome in relation to congenital malformations. J Intellect Disabil Res 1999;43:234-41. [PubMed]
- Han J, Lee Y, Yeom KH, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 2004;18:3016-27. [PubMed]
- Omran A, Elimam D, Webster KA, et al. MicroRNAs: a new piece in the paediatric cardiovascular disease puzzle. Cardiol Young 2013;23:642-55. [PubMed]
- Roberts A, Allanson J, Jadico SK, et al. The cardiofaciocutaneous syndrome. J Med Genet 2006;43:833-42. [PubMed]
- Perez E, Sullivan KE. Chromosome 22q11.2 deletion syndrome (DiGeorge and velocardiofacial syndromes). Curr Opin Pediatr 2002;14:678-83. [PubMed]
- Huang ZP, Chen JF, Regan JN, et al. Loss of microRNAs in neural crest leads to cardiovascular syndromes resembling human congenital heart defects. Arterioscler Thromb Vasc Biol 2010;30:2575-86. [PubMed]
- Veldtman GR, Connolly HM, Grogan M, et al. Outcomes of pregnancy in women with tetralogy of Fallot. J Am Coll Cardiol 2004;44:174-80. [PubMed]
- Burn J, Brennan P, Little J, et al. Recurrence risks in offspring of adults with major heart defects: results from first cohort of British collaborative study. Lancet 1998;351:311-6. [PubMed]
- Zellers TM, Driscoll DJ, Michels VV. Prevalence of significant congenital heart defects in children of parents with Fallot's tetralogy. Am J Cardiol 1990;65:523-6. [PubMed]
- Anderson RH, Baker EJ, Macartney FJ, et al. editors. Paediatric cardiology, 2nd edition. London: Churchill Livingstone, 2002;1213-502.
- Goldmuntz E, Clark BJ, Mitchell LE, et al. Frequency of 22q11 deletions in patients with conotruncal defects. J Am Coll Cardiol 1998;32:492-8. [PubMed]
- Bittel DC, Kibiryeva N, Marshall JA, et al. MicroRNA-421 Dysregulation is Associated with Tetralogy of Fallot. Cells 2014;3:713-23. [PubMed]
- Freeman RV, Otto CM. Spectrum of calcific aortic valve disease: pathogenesis, disease progression, and treatment strategies. Circulation 2005;111:3316-26. [PubMed]
- Sanford LP, Ormsby I, Gittenberger-de Groot AC, et al. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development 1997;124:2659-70. [PubMed]
- Bartram U, Molin DG, Wisse LJ, et al. Double-outlet right ventricle and overriding tricuspid valve reflect disturbances of looping, myocardialization, endocardial cushion differentiation, and apoptosis in TGF-beta(2)-knockout mice. Circulation 2001;103:2745-52. [PubMed]
- Jiao K, Langworthy M, Batts L, et al. Tgfbeta signaling is required for atrioventricular cushion mesenchyme remodeling during in vivo cardiac development. Development 2006;133:4585-93. [PubMed]
- Sridurongrit S, Larsson J, Schwartz R, et al. Signaling via the Tgf-beta type I receptor Alk5 in heart development. Dev Biol 2008;322:208-18. [PubMed]
- Charng MJ, Frenkel PA, Lin Q, et al. A constitutive mutation of ALK5 disrupts cardiac looping and morphogenesis in mice. Dev Biol 1998;199:72-9. [PubMed]
- Dean JC. Marfan syndrome: clinical diagnosis and management. Eur J Hum Genet 2007;15:724-33. [PubMed]
- Mizuguchi T, Matsumoto N. Recent progress in genetics of Marfan syndrome and Marfan-associated disorders. J Hum Genet 2007;52:1-12. [PubMed]
- Akutsu K, Morisaki H, Takeshita S, et al. Phenotypic heterogeneity of Marfan-like connective tissue disorders associated with mutations in the transforming growth factor-beta receptor genes. Circ J 2007;71:1305-9. [PubMed]
- Loeys BL, Chen J, Neptune ER, et al. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat Genet 2005;37:275-81. [PubMed]
- van de Laar IM, Oldenburg RA, Pals G, et al. Mutations in SMAD3 cause a syndromic form of aortic aneurysms and dissections with early-onset osteoarthritis. Nat Genet 2011;43:121-6. [PubMed]
- Lindsay ME, Schepers D, Bolar NA, et al. Loss-of-function mutations in TGFB2 cause a syndromic presentation of thoracic aortic aneurysm. Nat Genet 2012;44:922-7. [PubMed]
- Boileau C, Guo DC, Hanna N, et al. TGFB2 mutations cause familial thoracic aortic aneurysms and dissections associated with mild systemic features of Marfan syndrome. Nat Genet 2012;44:916-21. [PubMed]
- Peng Y, Song L, Zhao M, et al. Critical roles of miRNA-mediated regulation of TGFβ signalling during mouse cardiogenesis. Cardiovasc Res 2014;103:258-67. [PubMed]
- Saxena A, Tabin CJ. miRNA-processing enzyme Dicer is necessary for cardiac outflow tract alignment and chamber septation. Proc Natl Acad Sci U S A 2010;107:87-91. [PubMed]
- Chen JF, Murchison EP, Tang R, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A 2008;105:2111-6. [PubMed]
- Sussman MA, Völkers M, Fischer K, et al. Myocardial AKT: the omnipresent nexus. Physiol Rev 2011;91:1023-70. [PubMed]
- Rajan KS, Velmurugan G, Pandi G, et al. miRNA and piRNA mediated Akt pathway in heart: antisense expands to survive. Int J Biochem Cell Biol 2014;55:153-6. [PubMed]
- Pillai VB, Sundaresan NR, Gupta MP. Regulation of Akt signaling by sirtuins: its implication in cardiac hypertrophy and aging. Circ Res 2014;114:368-78. [PubMed]
- Liu J, van Mil A, Vrijsen K, et al. MiR-155 targets RIP1 and thus prevents necrotic cell death of human cardiomyocyte progenitor cells. J Cell Mol Med 2011;15:1474-82. [PubMed]
- Chen J, Zhang XD, Proud C. Dissecting the signaling pathways that mediate cancer in PTEN and LKB1 double-knockout mice. Sci Signal 2015;8:pe1. [PubMed]
- Jin Y, Chauhan SK, El Annan J, et al. A novel function for programmed death ligand-1 regulation of angiogenesis. Am J Pathol 2011;178:1922-9. [PubMed]
- Zhou M, Cai J, Tang Y, et al. MiR-17-92 cluster is a novel regulatory gene of cardiac ischemic/reperfusion injury. Med Hypotheses 2013;81:108-10. [PubMed]
- Yu QQ, Wu H, Huang X, et al. MiR-1 targets PIK3CA and inhibits tumorigenic properties of A549 cells. Biomed Pharmacother 2014;68:155-61. [PubMed]
- Chen T, Ding G, Jin Z, et al. Insulin ameliorates miR-1-induced injury in H9c2 cells under oxidative stress via Akt activation. Mol Cell Biochem 2012;369:167-74. [PubMed]
- Liu B, Wu X, Liu B, et al. MiR-26a enhances metastasis potential of lung cancer cells via AKT pathway by targeting PTEN. Biochim Biophys Acta 2012;1822:1692-704.
- Guo P, Nie Q, Lan J, et al. C-Myc negatively controls the tumor suppressor PTEN by upregulating miR-26a in glioblastoma multiforme cells. Biochem Biophys Res Commun 2013;441:186-90. [PubMed]
- Xu XD, Song XW, Li Q, et al. Attenuation of microRNA-22 derepressed PTEN to effectively protect rat cardiomyocytes from hypertrophy. J Cell Physiol 2012;227:1391-8. [PubMed]
- Nigam V, Sievers HH, Jensen BC, et al. Altered microRNAs in bicuspid aortic valve: a comparison between stenotic and insufficient valves. J Heart Valve Dis 2010;19:459-65. [PubMed]
- Cheng Y, Tan N, Yang J, et al. A translational study of circulating cell-free microRNA-1 in acute myocardial infarction. Clin Sci (Lond) 2010;119:87-95. [PubMed]
- Turchinovich A, Weiz L, Langheinz A, et al. Characterization of extracellular circulating microRNA. Nucleic Acids Res 2011;39:7223-33. [PubMed]
- Xing HJ, Li YJ, Ma QM, et al. Identification of microRNAs present in congenital heart disease associated copy number variants. Eur Rev Med Pharmacol Sci 2013;17:2114-20. [PubMed]



