
Characterization of microRNAs in spent culture medium associated with human embryo quality and development
Introduction
In vitro fertilization (IVF) is an effective and powerful assisted reproductive technology (ART), and has been commonly used to treat about 16% of couples who suffer from infertility worldwide (1). ARTs are well developed and have brought solutions to some fertility problems. IVF to produce embryos is advantageous because it bypasses challenges associated with fertilization, and offers an opportunity to better study early embryonic development (2). However, as a common means to measure embryo quality and potential of in vivo and IVF embryos in humans, morphological assessment often fails to indicate whether an embryo can successfully establish a pregnancy (3). Whereas embryos are often of similar morphology, the underlying genetics, such as transcriptomes, can be strikingly different. In addition, implantation failure after embryo transfer (ET) is also an obstacle to ARTs and has fettered the further improvement of the IVF success rate. Thus, a need exists for a better noninvasive method to efficiently select embryos based on their genetic potential and ability to establish a pregnancy.
MicroRNAs (miRNAs) are evolutionarily conserved single-stranded non-coding RNA molecules and important regulators in the post-transcriptional regulation of gene expression in various biological and physiological processes (4). miRNAs can regulate the levels of many target genes at the same time by partially complementary sequences and subsequent interference with messenger RNA (mRNA) stability and/or protein translation (4,5). Since they are stable in the extracellular environment, and their abnormal expressions are associated with physiological or pathological status, it is possible to treat miRNAs as new non-invasive biomarkers (6). It has also been widely reported that miRNAs were involved in embryo implantation, and associated with implantation failure (7,8). Kresowik et al. suggested that miR-31 plays a role in regulating the immune system during implantation (9), while Ioannidis et al. indicated that miR-26 might be a candidate biomarker for very early pregnancy in cows (10). miRNAs have attracted attention in several areas of medicine, including assisted reproduction.
An ideal approach to predict the implantation outcome should not compromise embryo implantation. It was believed that human embryos secrete miRNAs in easily collectable embryonic culture media, making it possible to use them as non-invasive biomarkers for embryo reproductive competence. The development of non-invasive biomarkers has driven extensive research in this area, with an overall aim to improve the success rate of implantation and IVF treatment. Abu-Halima et al. analyzed the miRNAs secreted from pre-implantation embryos into the embryonic culture media and identified that miR-634 was correlated with a positive pregnancy outcome (11). Then, in 2019, Abu-Halima and his colleagues reported that the abundance of miR-19b-3p was significantly lower in spent culture medium (SCM) samples associated with positive pregnancy (12). Fang et al. indicated that hsa-miR-26b-5p and hsa-miR-21-5p could serve as potential biomarkers for reproductive outcomes by detecting the miRNA profiles in the human embryo culture media (13).
In this study, we comprehensively analyzed the expression profile of miRNAs secreted by human pre-implantation embryos into SCM at different stages between a pregnant and non-pregnant group undergoing IVF treatment to investigate potential predictive markers of clinical outcomes of IVF-ET. By doing this, it would have a major impact on biomarker development and clinical practice for reproductive clinicians and scientists. Improved diagnosis of embryo implantation could have a profound effect on psychological and financial well-being on women and couples undergoing IVF treatment. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/atm-21-5029).
Methods
Embryo evaluation
Embryos at the cleavage on day 3 (D3 cleavage) and blastocyst stages on day 5 (D5 blastocyst) were evaluated using the ALPHA/ESHRE guidelines, and based on the number of blastomeres, rate of fragmentation, and symmetry. Cleavages with eight to 10 blastomeres, fragmentation <10%, or with mild unevenness, were considered of high-quality, and those with better morphology scores were maintained in culture until the blastocyst stage (at the 5th day of culture). Blastocysts that were scored as 4BB, 4BA, 4AB, and 4AA were regarded as good-quality blastocysts.
ET
In the case of a single blastocyst ET, only the most viable embryo was transferred on day 5. Serum or urinary ®-hCG was measured 14 days after ET, and its rise (>20 IU) across two consecutive blood tests or a positive urinary test indicated pregnancy. Clinical pregnancy was confirmed when a gestational sac with fetal heartbeat was revealed by ultrasound examination after 7 weeks of pregnancy.
SCM collection
The research described in the study was approved by the ethics committee of The Affiliated Drum Tower Hospital of Nanjing University Medical School (2019-198-01) and in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consent for use of their samples was provided by every participant. All embryos were obtained after controlled stimulation of the ovaries using gonadotropin-releasing hormone agonists and IVF techniques, and G1-PLUSTM/G2-PLUSTM (Vitrolife, Gothenburg, Sweden) was used for embryo culture. The SCM used for individual culture was collected individually from every embryo at D3 cleavage and D5 blastocyst during IVF cycles and stored at −80 °C until the analysis was performed.
RNA isolation and sequencing
Fifty-µL SCM from eight female/single-ETs, including three individuals in a non-pregnant group and five in a pregnant group were used for miRNA profiling. The media was collected from the same embryos on days 3 and 5. According to the manufacturer’s protocol, total RNA was extracted respectively from 16 SCM using miRNeasy Serum/Plasma Kit (Qiagen, Germany). Table S1 shows the characteristics of all these individuals. Small RNA sequencing was conducted by OE Biotech Co., Ltd. (Shanghai, China), while NEBNext Multiplex Small RNA Library Prep Set for Illumina was used to generate smRNA-seq libraries, with 20M reads per library obtained. The starting amount of input RNA was 1 ng. RNA sequencing was performed based on Illumina X-ten platform and the RNAs were aligned with bowtie and then subjected to the BLAST search against Rfam v.10.1 and GenBank databases. The known miRNAs were determined through aligning against miRBase v.21 database, and unannotated small RNAs were identified with mirdeep2 to predict novel miRNAs.
Identification of differentially expressed miRNAs (DEmiRNAs) and miRNA target prediction
The expression of miRNAs into SCM from the non-pregnant group was compared with those from the pregnant group. With DESeq, DEmiRNAs were determined with P value <0.05 and |log2FC| >1 between the groups at D3 cleavage and D5 blastocyst. The DEG algorithm was used to calculate P value in the R package. The targets of known DEmiRNAs were predicted using Miranda, with the parameter as follows: S ≥150, ΔG ≤−30 kcal/mol and demand strict 5’ seed pairing. Cytoscape soft (http://www.cytoscape.org/) was used to construct miRNA-mRNA interaction networks.
Functional enrichment analysis and protein-protein interaction (PPI) networks construction
CPDB (http://cpdb.molgen.mpg.de/CPDB) was applied to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of target genes of DEmiRNAs. GO enrichment analysis elucidated the functional annotations including biological process (BP), cellular component (CC), and molecular function (MF), and KEGG enrichment analysis was conducted to elucidate the target genes related signaling pathways, with the threshold set as P value <0.05. Online database STRING (https://string-db.org) was employed to analyze the PPI networks.
Quantitative real-time polymerase chain reaction (qRT-PCR) validation
In total, 15 individuals, including eight in the non-pregnant group and seven in the pregnant group, were recruited for qRT-PCR validation, and the media was collected from the same embryos on days 3 and 5. Total RNA extracted from 30 SCM with the miRNeasy Serum/Plasma Kit was used to validate the differential expression of miRNAs between the pregnant group and non-pregnant group by qRT-PCR analysis, with the qRT-PCR reactions performed in ABI 7300 Real-time PCR Detection System. All experiments were performed in three independent replicates. Relative gene expression was analyzed by 2−ΔΔCT method, and human hsa-U6 was used as endogenous controls for miRNA expression in analysis. The PCR primers are displayed in Table S2.
Validation in the Gene Expression Omnibus (GEO) database and receiver operating characteristic (ROC) analysis
GSE93810 was downloaded from the GEO database, which consists of 17 females with a positive pregnancy transfer and 39 females in whom no pregnancy occurred. The expression levels of selected DEmiRNAs were validated with GSE93810. Using pROC package in R language, we then performed the ROC analysis to assess the diagnostic value of DEmiRNAs. The area under the curve (AUC) was then calculated.
Statistical analysis
Statistical analyses of differential miRNAs between the non-pregnant group and the pregnant group were performed using R software. In addition, the GraphPad Prism Software, version 7.0 was used for the statistical analysis of experimental data. The results are expressed as means ± standard deviations (SDs). P<0.05 was considered to indicate a significant difference between the groups.
Results
Identification of DEmiRNAs
Compared with the pregnant group, 29 DEmiRNAs (11 up- and 18 down-regulated DEmiRNAs) were detected in the non-pregnant group at D3 cleavage with P value <0.05 and |log2FC| >1 (Figure 1A, Table 1), and 26 DEmiRNAs (19 up- and 7 down-regulated DEmiRNAs) were detected in the non-pregnant group at D5 blastocyst with P value <0.05 and |log2FC| >1 (Figure 1B, Table 2). In total, six known miRNAs, including hsa-miR-199a-3p>hsa-miR-199b-3p, hsa-miR-199a-5p, hsa-miR-379-5p, hsa-miR-432-5p, hsa-miR-99a-5p, and hsa-miR-483-5p, were identified in this analysis. The raw data have been deposited in the GEO database (GSE167961; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE167961).
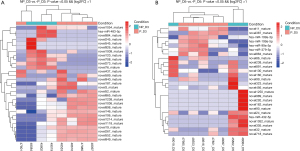
Table 1
| miRNA ID | Log2FC | P value | Regulation |
|---|---|---|---|
| hsa-miR-483-5p | Inf | 0.049484 | Up |
| novel1009_mature | 10.26512873 | 0.002501 | Up |
| novel1036_mature | −9.295460431 | 0.000136 | Down |
| novel1054_mature | 7.728030255 | 0.044975 | Up |
| novel106_mature | −6.64869016 | 0.017121 | Down |
| novel1118_mature | −8.863984044 | 0.00074 | Down |
| novel120_mature | 8.783172155 | 0.020206 | Up |
| novel1214_mature | −6.935777618 | 0.047552 | Down |
| novel1238_mature | −6.332283973 | 0.048518 | Down |
| novel146_mature | −5.16022631 | 0.000798 | Down |
| novel187_mature | −6.914511993 | 0.037222 | Down |
| novel19_mature | −4.166158994 | 0.00061 | Down |
| novel245_mature | Inf | 0.022999 | Up |
| novel267_mature | −8.143688147 | 0.004185 | Down |
| novel273_mature | 7.160261611 | 0.027341 | Up |
| novel3_mature | −8.846116416 | 0.007268 | Down |
| novel437_mature | −8.865099597 | 0.042348 | Down |
| novel52_mature>novel303_mature | −9.209703294 | 0.016312 | Down |
| novel545_mature>novel673_mature | Inf | 0.027824 | Up |
| novel552_mature | −7.264898066 | 0.000599 | Down |
| novel589_mature | −Inf | 0.020123 | Down |
| novel708_mature | 7.496888895 | 0.006455 | Up |
| novel76_mature | Inf | 0.000883 | Up |
| novel849_mature | −7.616105961 | 0.001598 | Down |
| novel865_mature | −9.598129639 | 0.007133 | Down |
| novel898_mature | −7.082397281 | 0.01306 | Down |
| novel926_mature | Inf | 0.027763 | Up |
| novel940_mature>novel986_mature | −7.790471729 | 0.020836 | Down |
| novel984_mature | Inf | 0.043655 | Up |
DEmiRNAs, differentially expressed miRNAs; SCM, spent culture medium; D3 cleavage, cleavage on day 3; miRNA, microRNA.
Table 2
| miRNA ID | Log2FC | P value | Regulation |
|---|---|---|---|
| hsa-miR-199a-3p>hsa-miR-199b-3p | −7.17811 | 0.026131 | Down |
| hsa-miR-199a-5p | −7.95985 | 0.038632 | Down |
| hsa-miR-379-5p | −7.50241 | 0.033988 | Down |
| hsa-miR-432-5p | Inf | 0.023522 | Up |
| hsa-miR-99a-5p | −4.93374 | 0.014704 | Down |
| novel1052_mature | Inf | 0.029945 | Up |
| novel11_mature>novel694_mature>novel887_mature | −7.96554 | 0.049573 | Down |
| novel1250_mature | Inf | 0.031455 | Up |
| novel130_mature | 9.108542 | 0.012543 | Up |
| novel160_mature | 10.96 | 0.001951 | Up |
| novel162_mature | Inf | 0.029479 | Up |
| novel238_mature | 9.675487 | 0.002768 | Up |
| novel24_mature | 7.646165 | 0.029516 | Up |
| novel292_mature | Inf | 0.023266 | Up |
| novel305_mature | Inf | 0.034748 | Up |
| novel323_mature>novel581_mature | Inf | 0.01271 | Up |
| novel32_mature | 8.267443 | 0.020619 | Up |
| novel387_mature | #NAME? | 0.014261 | Down |
| novel495_mature | 7.035155 | 0.038632 | Up |
| novel718_mature | 7.509777 | 0.019616 | Up |
| novel86_mature>novel741_mature | 9.724586 | 0.033219 | Up |
| novel884_mature | −8.42776 | 0.011719 | Down |
| novel891_mature | 11.00996 | 0.003768 | Up |
| novel90_mature>novel1213_mature | Inf | 0.029307 | Up |
| novel989_mature>novel1088_mature | Inf | 0.034597 | Up |
| novel99_mature | 8.797029 | 0.032628 | Up |
DEmiRNAs, differentially expressed miRNAs; SCM, spent culture medium; D5 blastocyst, blastocyst stages on day 5; miRNA, microRNA.
Construction of miRNA-mRNA interaction networks
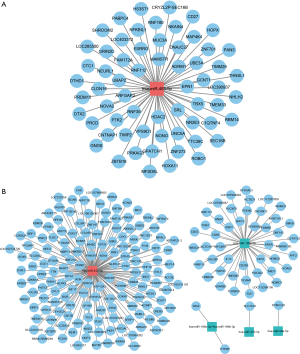
It has been reported that miRNAs can act by binding mRNA targets to repress their expression, allowing prediction of the targets of known DEmiRNAs. A total of 63 miRNA-mRNA pairs, involving one miRNA and 63 mRNAs, were obtained at D3 cleavage (Figure 2A), and a total of 203 miRNA-mRNA pairs, involving five miRNAs and 203 mRNAs, were obtained at D5 blastocyst (Figure 2B). In addition, we used TargetScan (http://www.targetscan.org/vert_72/) software to further validate the targeted relationship between miRNA and mRNA, such as hsa-miR-432-5p-ASPSCR1/ZNF384/GALNT9 and hsa-miR-483-5p-ZBTB16/HDAC2 (Figure S1).
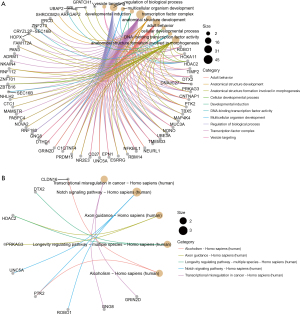
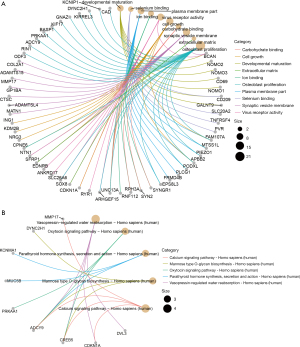
Functional enrichment analysis and PPI networks
The functional enrichment of the targets of known DEmiRNAs was performed to explore the function of the known miRNAs. The significant GO terms and pathways involved with genes targeted by miRNAs enriched in embryos at D3 cleavage and D5 blastocyst are displayed in Figures 3,4, respectively, and the results indicate these genes are associated with various BPs, including anatomical structure morphogenesis, cellular developmental process, cell communication, and cell signaling. With the STRING online database and Cytoscape software, the PPI networks of the targets of known DEmiRNAs were constructed, and as shown in Figure S2, the D3-specific PPI network included 27 proteins and the D5-specific PPI network included 92 proteins. Among them, HDAC2 (degree =5), ASPSCR1 (degree =4), ZNF384 (degree =4), GALNT9 (degree =4), and ZBTB16 (degree =3) were several hub proteins.
QRT-PCR confirmation and validation in the GEO database
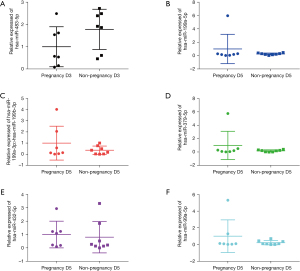
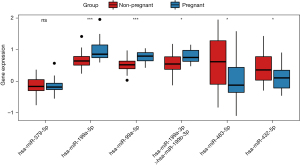
The expressions of six known miRNAs, including hsa-miR-199a-3p>hsa-miR-199b-3p, hsa-miR-199a-5p, hsa-miR-379-5p, hsa-miR-432-5p, hsa-miR-99a-5p, and hsa-miR-483-5p, were tested by qRT-PCR analysis and GEO database, and the qRT-PCR results indicated that except for hsa-miR-432-5p, the expression of the other miRNAs exhibited the same trend as the results of RNA sequencing (Figure 5). In addition, hsa-miR-199a-3p>hsa-miR-199b-3p, hsa-miR-199a-5p, hsa-miR-432-5p, hsa-miR-99a-5p, and hsa-miR-483-5p in GSE93810 exhibited the same trend as the results of RNA sequencing, which was statistically significant (Figure 6). ROC analysis indicated that hsa-miR-99a-5p (0.792) and hsa-miR-199a-5p (0.786) were with relatively high diagnostic value (Figure S3).
Discussion
Given the limitations of the low success rate of IVF caused by implantation failure after IVF-ET treatment, the investigation of biomarkers for embryo quality and predicting the outcomes of IVF-ET could contribute to the development of ARTs (14). Recently, the role of a variety of biomarkers including miRNAs, such as proteomics and hormones in SCM, has been extensively studied. Borges et al. suggested that miR-142-3p may be a potential biomarker of blastocyst implantation failure (15), and increased levels of apolipoprotein A1 were present in SCM containing blastocysts of higher morphologic grade, which may suggest a role for lipoproteins in early embryologic development (16). Chen et al. indicated secreted beta-human chorionic gonadotropin (HCG) as a useful biomarker for embryo selection in IVF-ET procedure (17), while Wang et al. reported that the concentration of HCG in SCM is positively correlated with the status of early embryo development and implantation rate, thus serves as a useful marker for embryo selection in the IVF-ET procedure (18). We analyzed the profiles of miRNAs secreted from pre-implantation embryos into SCM between a pregnant and non-pregnant group undergoing IVF treatment as biomarkers to predict the outcomes of IVF-ET. A total of six known miRNAs were identified in SCM of the pregnant group, and among them, hsa-miR-199a-5p, hsa-miR-483-5p, and hsa-miR-432-5p were three miRNAs with abundant target genes.
In this study, hsa-miR-199a-5p was detected to present an increased trend in SCM of the pregnant group. Considerable evidence has demonstrated that miR-199a-5p was involved in various cancers (19), and mmu-miR-199a-5p was reported to silence the self-renewal of mouse embryonic stem cells (20), and Tan et al. reported that, compared to an in vivo-fertilized group, down-regulated miR-199a-5p in IVF blastocysts was responsible for the lower developmental potential and subsequent viability (19). Li et al. suggested that ssc-miR-199a-5p may play a crucial role for implantation by investigating miRNAome of the endometrium of pregnancy and non-pregnancy groups in pigs (21). Compared with fresh mouse blastocysts, up-regulated mmu-miR-199a-5p was detected in the vitrified blastocysts, suggesting vitrification, a method commonly used in the cryopreservation of mammalian blastocysts, may decrease the implantation potential of vitrified blastocysts (22). In summary, we conclude hsa-miR-199a-5p is closely related to embryo implantation, but the underlying mechanism of hsa-miR-199a-5p in perimplantation embryos requires further exploration.
As over-expressed miR-483-5p suppressed glioma cell proliferation and induced a G0/G1 arrest, it is considered to be a tumor suppressor (23). It was reported that miR-483-5p regulated the progesterone concentration in a steroidogenic human granulosa-like tumor cell line (24). Down-regulated miR-483-5p was observed in cumulus cells of metaphase II oocytes from women with polycystic ovary syndrome (25), and an exploratory study uncovered that dysregulated miR-483-5p was associated with pregnancy-related adverse outcomes (26). It has been reported that miR-483-5p was a human-specific regulator of MeCP2 levels in human fetal brains (27), while decreased miR-483-5p was detected in placentas from patients with preeclampsia and preterm labor as compared to normal term pregnancies, indicating which patient’s altered expression and function may associate with pregnancy complications (28). It has been reported that ZBTB16 is essential for decidualization, which supports embryo implantation and placentation, as well as subsequent events that together ensure a successful pregnancy (29), and that HDAC2 was poorly expressed in placental tissue samples from pregnant women with preeclampsia (30,31). In this study, down-regulated hsa-miR-483-5p was detected in SCM of a pregnant group and ZBTB16/HDAC2 were targets of hsa-miR-483-5p. These findings may indicate that hsa-miR-483-5p plays a unique role via regulating ZBTB16/HDAC2 in early embryo development.
In recent years, the role of miR-432-5p in cancers has been extensively studied. A previous study revealed that decreased miR-432-5p was reported to promote breast cancer cell growth and migration by targeting E2F3 (32). It has also been reported that miR-432-5p was implicated in facilitating cell progression by activating Wnt/β-catenin signaling and promoting cell growth and metastasis in human hepatocellular carcinoma (33,34). Yang et al. reported that lower levels of miR-432-5p were observed in both glioma tissues and cells (35), and it has been extensively reported that translocation t(X;17)(p11;q25) of ASPSCR1 with transcription factor TFE3 gene results in a ASPSCR1-TFE3 fusion protein in alveolar soft part sarcoma and in renal cell carcinomas (36,37). ZNF384-related fusion genes were detected in acute lymphoblastic leukemia (38), and GALNT9 was demonstrated to be well conserved in zebrafish during embryonic development (39). To the best of our knowledge, there is no study linking miR-432-5p and ASPSCR1/ZNF384/GALNT9 to embryo development. In the current study, the expression of hsa-miR-432-5p was reduced in SCM of the pregnant group, and ASPSCR1/ZNF384/GALNT9 were targets of hsa-miR-432-5p, which indicates the role of hsa-miR-432-5p-ASPSCR1/ZNF384/GALNT9 in embryo development should be studied further.
Firstly, improving embryo quality and selecting high-quality embryos for transfer is a critical step to improve the success rate of embryo implantation. Secondly, couples should make lifestyle adjustments, including quitting smoking and alcohol, and try to maintain a normal body mass index (BMI) by gaining or losing weight. It has been suggested that extending embryo culture to day 5 or 6 in order to transfer the embryo at the blastocyst stage increases the implantation rate (40). In addition, assisted hatching has been proposed as one technique to improve implantation and pregnancy rates following IVF Improving embryo quality and selection (41).
Conclusions
In conclusion, the three miRNAs mentioned above may be involved in early embryo development by regulating abundant target genes. These findings indicate the possibility of using miRNAs secreted by embryos into SCM as non-invasive biomarkers to predict the outcomes of IVF. Our present study lays the foundation for developing biomarkers for predicting embryo quality and exploring the roles of embryonic miRNAs found in SCM. The limitations of present study are the low abundance of miRNAs, small sample size, and lack of experimental verification. Although the results of qRT-PCR and validation in the GEO database suggested the reliability of our RNA-sequencing results, ex-vivo experiments are required to confirm the role of miRNAs identified in this study, and clinical validation with larger sample sizes is mandatory to confirm its effectiveness.
Acknowledgments
Funding: This study was supported by the Fertility Research Program of Young and Middle-Aged Physicians from China Health Promotion Foundation, Medical Scientific Research Project of Jiangsu Provincial Health Commission (No. H2019011), Special Fund for Clinical Medical Research of Chinese Medical Association (No. 17020450714), Clinical Research Special Fund Project of the Affiliated Drum Tower Hospital of Nanjing University Medical School (Cultivation Project) (No. 2021-LCYJ-PY-08), Fertility Research Program of Young and Middle-Aged Physicians from China Health Promotion Foundation, 2021, and General Project of Nanjing Medical Science and Technology Development (No. YKK20064).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-5029
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-5029
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-5029). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The research described in the study was approved by the ethics committee of The Affiliated Drum Tower Hospital of Nanjing University Medical School (2019-198-01) and in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consent for use of their samples was provided by every participant.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Frederiksen Y, Farver-Vestergaard I, Skovgård NG, et al. Efficacy of psychosocial interventions for psychological and pregnancy outcomes in infertile women and men: a systematic review and meta-analysis. BMJ Open 2015;5:e006592 [Crossref] [PubMed]
- Kropp J, Khatib H. Characterization of microRNA in bovine in vitro culture media associated with embryo quality and development. J Dairy Sci 2015;98:6552-63. [Crossref] [PubMed]
- Van Soom A, Mateusen B, Leroy J, et al. Assessment of mammalian embryo quality: what can we learn from embryo morphology? Reprod Biomed Online 2003;7:664-70. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005;433:769-73. [Crossref] [PubMed]
- Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513-8. [Crossref] [PubMed]
- Liu W, Niu Z, Li Q, et al. MicroRNA and Embryo Implantation. Am J Reprod Immunol 2016;75:263-71. [Crossref] [PubMed]
- Liang J, Wang S, Wang Z. Role of microRNAs in embryo implantation. Reprod Biol Endocrinol 2017;15:90. [Crossref] [PubMed]
- Kresowik JD, Devor EJ, Van Voorhis BJ, et al. MicroRNA-31 is significantly elevated in both human endometrium and serum during the window of implantation: a potential biomarker for optimum receptivity. Biol Reprod 2014;91:17. [Crossref] [PubMed]
- Ioannidis J, Donadeu FX. Circulating miRNA signatures of early pregnancy in cattle. BMC Genomics 2016;17:184. [Crossref] [PubMed]
- Abu-Halima M, Häusler S, Backes C, et al. Micro-ribonucleic acids and extracellular vesicles repertoire in the spent culture media is altered in women undergoing In Vitro Fertilization. Sci Rep 2017;7:13525. [Crossref] [PubMed]
- Abu-Halima M, Khaizaran ZA, Ayesh BM, et al. MicroRNAs in combined spent culture media and sperm are associated with embryo quality and pregnancy outcome. Fertil Steril 2020;113:970-80.e2. [Crossref] [PubMed]
- Fang F, Li Z, Yu J, et al. MicroRNAs secreted by human embryos could be potential biomarkers for clinical outcomes of assisted reproductive technology. J Adv Res 2021;31:25-34. [Crossref] [PubMed]
- Norwitz ER, Schust DJ, Fisher SJ. Implantation and the survival of early pregnancy. N Engl J Med 2001;345:1400-8. [Crossref] [PubMed]
- Borges E Jr, Setti AS, Braga DP, et al. miR-142-3p as a biomarker of blastocyst implantation failure - A pilot study. JBRA Assist Reprod 2016;20:200-5. [Crossref] [PubMed]
- Mains LM, Christenson L, Yang B, et al. Identification of apolipoprotein A1 in the human embryonic secretome. Fertil Steril 2011;96:422-7.e2. [Crossref] [PubMed]
- Xiao-Yan C, Jie L, Dang J, et al. A highly sensitive electrochemiluminescence immunoassay for detecting human embryonic human chorionic gonadotropin in spent embryo culture media during IVF-ET cycle. J Assist Reprod Genet 2013;30:377-82. [Crossref] [PubMed]
- Wang H, Zhang R, Han D, et al. Association of human chorionic gonadotropin level in embryo culture media with early embryo development. Nan Fang Yi Ke Da Xue Xue Bao 2014;34:1039-41, 1047. [PubMed]
- Tan K, Wang X, Zhang Z, et al. Downregulation of miR-199a-5p Disrupts the Developmental Potential of In Vitro-Fertilized Mouse Blastocysts. Biol Reprod 2016;95:54. [Crossref] [PubMed]
- Wang Y, Melton C, Li YP, et al. miR-294/miR-302 promotes proliferation, suppresses G1-S restriction point, and inhibits ESC differentiation through separable mechanisms. Cell Rep 2013;4:99-109. [Crossref] [PubMed]
- Li W, Xi Y, Xue S, et al. Sequence analysis of microRNAs during pre-implantation between Meishan and Yorkshire pigs. Gene 2018;646:20-7. [Crossref] [PubMed]
- Zhao X, Hao H, Du W, et al. Effect of vitrification on the microRNA transcriptome in mouse blastocysts. PLoS One 2015;10:e0123451 [Crossref] [PubMed]
- Wang L, Shi M, Hou S, et al. MiR-483-5p suppresses the proliferation of glioma cells via directly targeting ERK1. FEBS Lett 2012;586:1312-7. [Crossref] [PubMed]
- Sang Q, Yao Z, Wang H, et al. Identification of microRNAs in human follicular fluid: characterization of microRNAs that govern steroidogenesis in vitro and are associated with polycystic ovary syndrome in vivo. J Clin Endocrinol Metab 2013;98:3068-79. [Crossref] [PubMed]
- Shi L, Liu S, Zhao W, et al. miR-483-5p and miR-486-5p are down-regulated in cumulus cells of metaphase II oocytes from women with polycystic ovary syndrome. Reprod Biomed Online 2015;31:565-72. [Crossref] [PubMed]
- Rodosthenous RS, Burris HH, Sanders AP, et al. Second trimester extracellular microRNAs in maternal blood and fetal growth: An exploratory study. Epigenetics 2017;12:804-10. [Crossref] [PubMed]
- Han K, Gennarino VA, Lee Y, et al. Human-specific regulation of MeCP2 levels in fetal brains by microRNA miR-483-5p. Genes Dev 2013;27:485-90. [Crossref] [PubMed]
- Mayor-Lynn K, Toloubeydokhti T, Cruz AC, et al. Expression profile of microRNAs and mRNAs in human placentas from pregnancies complicated by preeclampsia and preterm labor. Reprod Sci 2011;18:46-56. [Crossref] [PubMed]
- Katoh N, Kuroda K, Tomikawa J, et al. Reciprocal changes of H3K27ac and H3K27me3 at the promoter regions of the critical genes for endometrial decidualization. Epigenomics 2018;10:1243-57. [Crossref] [PubMed]
- Gao Y, She R, Wang Q, et al. Up-regulation of miR-299 suppressed the invasion and migration of HTR-8/SVneo trophoblast cells partly via targeting HDAC2 in pre-eclampsia. Biomed Pharmacother 2018;97:1222-8. [Crossref] [PubMed]
- Fan Y, Dong Z, Zhou G, et al. Elevated miR-23a impairs trophoblast migration and invasiveness through HDAC2 inhibition and NF-κB activation. Life Sci 2020;261:118358 [Crossref] [PubMed]
- Liu Y, Lu C, Zhou Y, et al. Circular RNA hsa_circ_0008039 promotes breast cancer cell proliferation and migration by regulating miR-432-5p/E2F3 axis. Biochem Biophys Res Commun 2018;502:358-63. [Crossref] [PubMed]
- Jiang N, Chen WJ, Zhang JW, et al. Downregulation of miR-432 activates Wnt/β-catenin signaling and promotes human hepatocellular carcinoma proliferation. Oncotarget 2015;6:7866-79. [Crossref] [PubMed]
- Liu H, Xue L, Song C, et al. Overexpression of circular RNA circ_001569 indicates poor prognosis in hepatocellular carcinoma and promotes cell growth and metastasis by sponging miR-411-5p and miR-432-5p. Biochem Biophys Res Commun 2018;503:2659-65. [Crossref] [PubMed]
- Yang G, Han B, Feng T. ZFAS1 knockdown inhibits viability and enhances cisplatin cytotoxicity by up-regulating miR-432-5p in glioma cells. Basic Clin Pharmacol Toxicol 2019;125:518-26. [Crossref] [PubMed]
- Kauffman EC, Ricketts CJ, Rais-Bahrami S, et al. Molecular genetics and cellular features of TFE3 and TFEB fusion kidney cancers. Nat Rev Urol 2014;11:465-75. [Crossref] [PubMed]
- Paoluzzi L, Maki RG. Diagnosis, Prognosis, and Treatment of Alveolar Soft-Part Sarcoma: A Review. JAMA Oncol 2019;5:254-60. [Crossref] [PubMed]
- Hirabayashi S, Ohki K, Nakabayashi K, et al. ZNF384-related fusion genes define a subgroup of childhood B-cell precursor acute lymphoblastic leukemia with a characteristic immunotype. Haematologica 2017;102:118-29. [Crossref] [PubMed]
- Nakayama Y, Nakamura N, Kawai T, et al. Identification and expression analysis of zebrafish polypeptide α-N-acetylgalactosaminyltransferase Y-subfamily genes during embryonic development. Gene Expr Patterns 2014;16:1-7. [Crossref] [PubMed]
- Coughlan C, Ledger W, Wang Q, et al. Recurrent implantation failure: definition and management. Reprod Biomed Online 2014;28:14-38. [Crossref] [PubMed]
- Practice Committee of Society for Assisted Reproductive Technology. The role of assisted hatching in in vitro fertilization: a review of the literature. A Committee opinion. Fertil Steril 2008;90:S196-8. [Crossref] [PubMed]


