
LncRNA SNHG16 promotes Schwann cell proliferation and migration to repair sciatic nerve injury
Introduction
Peripheral nerve injury is common (1), and its repair and regeneration is a physiological process involving multiple factors (2). Increasing attention is being paid to nerve injury and repair, especially its molecular mechanism (3), which has been found to be regulated by many molecules and cells. After injury, Schwann cells (SCs) divide, proliferate, and secrete a variety of neurotrophic factors providing a microenvironment suitable for repair and regeneration (4,5).
With a length of more than 200 nucleotides (6), long non-coding RNAs (lncRNAs) the largest class of non-coding RNAs, and compared with mRNA encoding protein, have stronger tissue and cell expression specificity (7). Recent studies have found many lncRNAs are differentially expressed after peripheral nerve injury and play an important role in peripheral nerve regeneration (8-10). LncRNA UC.217 was down-regulated in dorsal root ganglion neurons after sciatic nerve injury, while its elimination significantly promoted the growth of cultured dorsal root ganglion neurons (11). The down-regulation of lncRNA BC089918 also promoted neurite growth in dorsal root ganglia, and the co-expression network showed that its potential targets included FAM57B, KCNS1, and CACNG2. Among these three targets, KCNS1 was expressed in sensory neurons, and its protein level was significantly down-regulated after sciatic nerve injury, affecting neuronal excitability (12). Snorna hostgene16 (SNHG16) is a lncRNA located on human chromosome 17 17q25.1 (13,14). Existing studies have reported that SNHG16 expression is up-regulated in patients with neuroblastoma (15), and its expression is correlated with bladder cancer invasion (16), with cell proliferation and apoptosis in colon cancer (17). However, the role of SNHG16 in sciatic nerve injury has not been reported.
The purpose of this study was to determine whether SNHG16 could be used as a gene to promote nerve repair by examining the effects of its overexpression on the proliferation, migration, and invasion of SCs during sciatic nerve injury. Further analysis of the molecular mechanism was also undertaken to confirm whether SNHG16 could up-regulate signal activator of transcription 3 (STAT3) expression through endogenous competitive sponge adsorption of miR-93-5p.
We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-3971).
Methods
Sciatic nerve injury model
Forty healthy adult SPF SD rats were purchased from the Beijing Vital River Laboratory Animal Technology Co., Ltd. and randomly divided into 0, 1, 4, and 7 d groups, 5 in each group. To establish a sciatic nerve injury model, rats were anesthetized by intraperitoneal injection of 2% pentobarbital (40 mg/kg), and after prone fixation, a longitudinal incision (about 1.5 cm) was made in the posterior femoral section of the left lower limb under aseptic conditions. The left sciatic nerve was exposed, then cut laterally 8 mm from the lower margin of the piriformis muscle. The epineurium was then sutured with a “10-0” micro-noninvasive suture in accordance with the vascular direction on the epineurium surface, and the incision closed layer by layer with thicker sutures. Cage feeding after operation to promote animal activities and prevent plantar ulcers was implemented. The rats were killed by decapitation. The corresponding author was aware of the group allocation at the different stages of the experiment (during the allocation, the conduct of the experiment, the outcome assessment, and the data analysis). Experiments were performed under a project license (No.: 2020-012) granted by ethics board of the Second Affiliated Hospital of Soochow University, in compliance with USA Institute for Laboratory Animal Research (ILAR) Guide for the care and use of animals. A protocol was prepared before the study without registration.
Isolation and culture of Schwann cell
Five rats were sacrificed after cervical dislocation and about 1.5 cm of bilateral sciatic nerve was taken for SC culture by the enzyme digestion method (18). The general procedure was as follows: the epineurium was carefully peeled off under dissection microscope and the nerve was cut into pieces with a volume of about 1 mm3. Type II collagenase was added at a concentration of 0.1% and the volume was about 20 times that of nerve tissue. After fully mixed digestion in a carbon dioxide incubator for 35–45 min and there was no obvious tissue mass, the supernatant was discarded after centrifugation at 1,000 r/min for 5 min. DMEM/F12 medium containing 10% fetal bovine serum was added to resuspend the cells, which were then gently beaten until thoroughly mixed. Cells were inoculated in a 25 cm2 cell culture flask coated with laminin and the density was (3.1±0.17)×105 cells/mL. SCs were purified by low concentration collagenase differential digestion (19).
Cell transfection
Transfection was performed using Lipofectamine 2000 Liposome Transfection according to the manufacturer’s instructions. 0.5 µg transgenic plasmid was mixed with 50 µL incomplete medium DMEM (Liquid A), and 1 µL Lipofectamine 2000 was mixed with 50µL incomplete medium DMEM (Liquid B). Liquid A and liquid B were then mixed and incubated at room temperature for 20 min to form a liposome complex, to which 0.8 mL incomplete medium DMEM was added before being placed on cell culture plates. These were then incubated at 37 °C in an incubator containing 5% CO2 for 6h, and the complete DMEM was replaced and cultured for 48 hours. Each group had five duplicate holes.
QRT-PCR
Total RNA was extracted by the Trizol method (Sigma Corporation, USA) and cDNA was obtained by reverse transcription. The reaction conditions were 37 °C for 15 min and 85 °C for 15 s. The reaction system consisted of a 2 µL 5× reaction mixture with 1 µL RNA and 7 µL H2O. The primer sequence was as follows: SNHG16 upstream 5'-CCGGTTTTCCAGTTCTTGCA-3', and downstream 5'-GCCTCACAGGGAAACTTCATG-3'. The internal reference of GAPDH was upstream 5'-CaAGGTCATCCATGACAACTTTG-3' and downstream 5'-GTCCACCACCCT⁃GTTGCTGTAG-3'. PCR was performed using cDNA as the template. The reaction system consisted of 10 µL SYBR, 1 µL cDNA, 1 µL primer, and 7 µL H2O, and the reaction conditions were 95 °C for 10 min, 95 °C for 30 s, 60 °C for 15 s, 72 °C for 20 s (40 cycles), and 72 °C for 10 min. The relative expression level was calculated by the 2−CT method.
Fluorescence in situ hybridization
SCs fixed with paraformaldehyde were treated with 0.1%Triton-X 100 for 5 min to improve the permeability of the probe, then fixed with 4% paraformaldehyde for 15 min and rinsed with PBSM (0.01M PBS containing 5 mM MgCl2). 50% formamide/2× SSC was used for equilibrium at room temperature for 10 min. A 100 µL fluorescent probe solution (concentration 2.5 µmol/L) was then added, and the reaction was carried out at 50% formamide/2× SSC 37 °C for 3 h, then flushed with PBSM. Glycerin was sealed after dehydration with gradient alcohol, then a fluorescence microscope was used for observation.
CCK-8
U251 cells in logarithmic growth phase were seeded into 96-well plates at a density of 1×104 cells/well, 100 µL per well. After cells adhered to the wall, 10 µL CCK-8 solutions were added for 72 h. The optical density (OD) was measured by a microplate analyzer to reflect the proliferation ability of cells.
EdU experiment
4×103 cells per well were seeded into 96-well plates and cultured 48 h after transfection. The EdU solution was diluted with cell medium at a ratio of 1,000:1, and after culture for 4 h, the cells were washed twice with PBS. Samples were fixed with 4% methanol and stained with Apollo for 5 min each time and DAPI was used to dye the nucleus. Multi-field photos were taken under the microscope for counting.
Transwell experiment
The upper chamber of the Transwell plate was first coated with matrix adhesive, and the logarithmic growth phase cells of each group were then seeded into the 24-well plates of the Transwell compartment. Cells were added into the upper chamber at a density of 2×105 cells/mL (100 µL per well), and a medium containing fetal bovine serum (250 µL/well) was added to the lower chamber. After 48 h of culture, the compartment was removed and the cells in the suprapore compartment were swabbed with a cotton swab. The upper and lower sides of the chamber were flushed twice with phosphate buffer and cells attached to the microporous membrane of the inferior compartment were then fixed with paraformaldehyde. Cells were then stained with crystal violet for 15 min, and after drying, were observed under microscope (400×) with five fields of vision used for counting.
Scratch test
SCs were collected after transfection, digested with trypsin solution for 1 min, then centrifugal resuspended and inoculated into 6-well plates at a concentration of 2×105 cells/mL. After the cells were fully integrated and lined the bottom of the culture plate, they were scratched with the 100 µL tip of the spear. The culture was continued for 48 hours, and a microscope used to take photos which allowed cell mobility to be calculated.
Double luciferase reporter gene assay
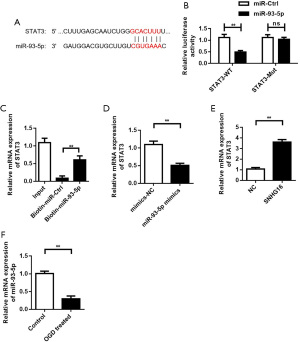
Bioinformatics software TargetScan was used to predict the binding sites of miR-93-5p and lncRNA SNHG16 3'UTR, and the gene fragment of the binding site was amplified by qRT-PCR. The segment was inserted into the luciferase reporter gene vector (pmirglo-REPORT, pMIR-report) and SNHG16 wild plasmid constructed. The SNHG16 mutant plasmid was then constructed by a gene site image technique. According to the instructions of Lipofectamine 3000, the wild plasmid, the mutant plasmid, and the internal luciferase plasmid PRL-thymidine kinase (TK) were co-transfected into the cells, and after incubation for 48h, the luciferase activity was detected by the luciferase activity kit. Relative luciferase activity was expressed by the ratio of luciferase activity of firefly to luciferase activity of Renilla Luciferase.
RNA pull down experiment
Biotin-labeled miR-93-5p was transfected into the cells after their density was more than 80%. After 36 h of culture, the medium was discarded, the cells were washed twice with PBS, and RNA pull down lysis reagent was added before collection. 50 µL of pyrolysis solution was absorbed for input detection, while M280 streptomycin beads were coated with yeast tRNA 60 min in advance. M280 streptomycin magnetic beads were added to the sample solution before incubation at 4 °C for 3 h. The supernatant was then discarded, and the cracking solution was added and washed twice. The sample was then washed with low salt solution and high salt solution twice, respectively, before the supernatant was removed and Trizol added. RNA was extracted and qRT-PCR was performed to verify the content of target genes captured by the miR-93-5p probe.
Statistical analysis
SPSS18.0 software was used for data statistical analysis. Measurement data were expressed as mean ± standard deviation and the t-test was used for comparison between the two groups. One-way analysis of variance was used for comparison between groups and P<0.05 or P<0.01 indicated a statistically significant difference.
Results
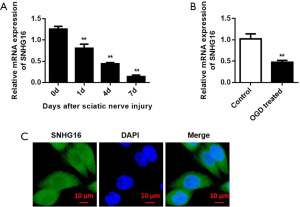
Expression of lncRNA SNHG16 decreased after sciatic nerve injury.
To investigate the role of SNHG16 in sciatic nerve injury, we constructed nerve injury models in vivo and in vitro. The expression of SNHG16 was detected by qRT-PCR at 0, 1, 4, and 7 days after injury (n=10), and the results showed that the expression of SNHG16 decreased gradually with the extension of time (Figure 1A). In addition, qRT-PCR was used to detect the expression of SNHG16 in primary cultured SCs after OGD treatment. The cells were hypoxic under the condition of low oxygen and high nitrogen in a three-gas incubator, and a medium with low sugar and no serum was used. The experimental results showed that compared with the control group, the expression of SNHG16 in SCs treated with OGD was decreased, with statistical significance (Figure 1B). Subsequently, the cell sublocalization of SNHG16 was detected by fluorescence in situ hybridization (FISH) and the localization of the SNHG16 fluorescent probe in cells was captured by fluorescence microscopy. The results of the FISH experiment showed that SNHG16 was localized in the cytoplasm (Figure 1C).
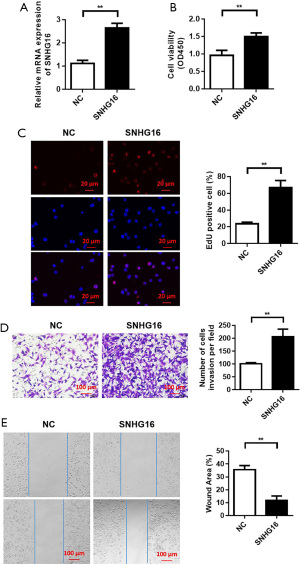
Overexpression of SNHG16 promoted the proliferation, migration, and invasion of SCs
QRT‐PCR was used to determine the transfection efficiency of overexpressed SNHG16. PCR results showed that the expression of SNHG16 in the SNHG16 group was significantly higher than in the control group, indicating the SC model with overexpression of SNHG16 was successfully constructed (Figure 2A). CCK8 results showed that with the increase of culture time, the proliferation ability of SCs in the overexpressed SNHG16 group was significantly enhanced compared with the control group, and the difference was statistically significant (Figure 2B). After transfection with SNHG16, SC proliferation was analyzed by EdU staining, and the results showed that the number of EdU positive SCs in the overexpressed SNHG16 group was significantly higher than in the control group, and the difference was statistically significant (Figure 2C). Collectively, these results suggest the overexpression of SNHG16 could promote the proliferation of SCs. The invasion ability of SCs transfected with SNHG16 was detected by Transwell and the experimental results showed that its overexpression promoted the invasion of SCs compared to the control group (Figure 2D). Similarly, the migration ability of SCs transfected with SNHG16 was detected by the scratch test, and experimental results showed that overexpression of SNHG16 promoted SC migration compared to the control group (Figure 2E).
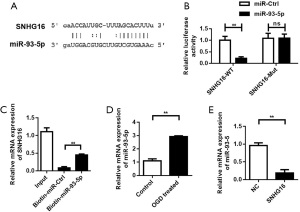
SNHG16 plays a biological role as a “sponge” of miR-93-5p
To study the downstream regulatory molecular mechanism of SNHG16, we predicted the binding sites of SNHG16 and miR-93-5p through Starbase (Figure 3A) and verified the binding reliability of miR-93-5p with SNHG16 through dual luciferase reporter gene assay. While the results showed that in the wild-type vector, overexpression of miR-93-5p could significantly reduce the luciferase activity of SNHG16, the inhibitory effect of miR-93-5p on luciferase activity was lost after co-transfection of the vector with miR-93-5p and mutation at the target site (Figure 3B). In addition, we verified the binding of SNHG16 with miR-93-5p through a miRNA pull down experiment, and the qRT-PCR results showed that Biotin-miR-93-5p was significantly enriched in SNHG16 (Figure 3C). The detection results of miR-93-5p expression in the SC OGD model showed that, compared with the control group, the expression of miR-93-5p was up-regulated (Figure 3D). PCR results showed that the mRNA expression level of miR-93-5p in SCs in the SNHG16 overexpression group was significantly decreased compared with the control group (Figure 3E), indicating the overexpression of SNHG16 inhibited the expression of miR-93-5p.
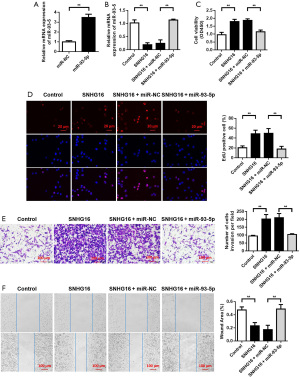
Overexpression of miR-93-5p reversed the promoting effects of SNHG16 on SC proliferation and invasion
In SCs, we transfected blank plasmid, SNHG16 overexpressed plasmid, co-transfected SNHG16 overexpressed plasmid, and miR-NC, and co-transfected SNHG16 overexpressed plasmid and miR-93-5p mimics. We first detected the transfection efficiency of miR-93-5p by qRT-PCR and the results showed miR-93-5p mimics could up-regulate the expression of miR-93-5p (Figure 4A). The detection results of miR-93-5p expression after different treatments showed that, compared with cells transfected with blank plasmid, the expression of miR-93-5p in cells transfected with SNHG16 overexpressed plasmid decreased. Further, compared with cells co-transfected with SNHG16 overexpressing plasmid and miR-NC, the expression of miR-93-5p in those co-transfected with SNHG16 overexpressing plasmid and miR-93-5p mimics increased (Figure 4B). CCK-8 was used to detect the changes of cell proliferation, and the results showed that, compared with cells transfected with blank plasmid, the proliferation ability of those transfected with SNHG16 overexpression plasmid was enhanced. In addition, compared with cells co-transfected with SNHG16 overexpression plasmid and miR-NC, the proliferation ability of cells co-transfected with SNHG16 overexpression plasmid and miR-93-5p mimics was decreased (Figure 4C,4D). We also used Transwell assay and scratch assay to detect changes in cell invasion and migration and the results showed that compared with cells transfected with blank plasmid, the invasion and migration ability of cells transfected with SNHG16 overexpression plasmid was enhanced. However, compared with cells co-transfected with SNHG16 overexpressed plasmid and miR-NC, the invasion and migration ability of cells co-transfected with SNHG16 overexpressed plasmid and miR-93-5p mimics were decreased (Figure 4E,4F).
STAT3 was the target gene of miR-93-5p
By binding with miR-93-5p, SNHG16 may affect the binding of miR-93-5p to its target genes. We predicted the target gene of miR-93-5p and found that STAT3 could be a target gene of miR-93-5p (Figure 5A). The question of whether miR-93-5p could bind to STAT3 was explored with dual luciferase reporter gene assay and showed that in wild-type vectors, overexpression of miR-93-5p significantly reduced the activity of STAT3 luciferase. However, the inhibitory effect of miR-93-5p on luciferase activity was lost after co-transfection of the vector with miR-93-5p and mutation at the target site (Figure 5B). The binding of miR-93-5p with STAT3 was also verified by miRNA pull down (Figure 5C). We detected the effect of miR-93-5p on the effective expression level of STAT3 in SC lines by PCR, and the results showed that overexpression of miR-93-5p significantly reduced the effective expression of STAT3 (Figure 5D). PCR results showed that compared with the control group, the mRNA expression level of STAT3 in SCs in the SNHG16 overexpression group was significantly increased (Figure 5E), showing overexpression of SNHG16 promoted the expression of STAT3. In addition, STAT3 expression was down-regulated in the SC OGD model (Figure 5F).
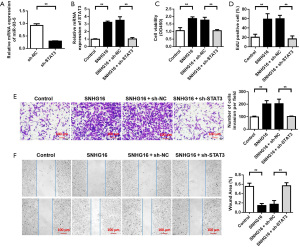
Stat 3 knockdown reversed the proliferation and invasion promotion effect of SNHG16 on SCs
After elucidating the ceRNA mechanism of SNHG16/miR-93-5p/STAT3 in SCs, we further studied the effect of STAT3 knockdown on SNHG16 through rescue experiments. QRT-PCR results showed that sh-STAT3 plasmid could reduce STAT3 expression (Figure 6A), and further results showed that the expression of STAT3 in cells transfected with SNHG16 overexpressing plasmid was increased compared with that in cells transfected with blank plasmid. Compared with cells co-transfected with SNHG16 overexpressing plasmid and sh-NC, the expression of STAT3 in cells co-transfected with SNHG16 overexpressing plasmid and sh-STAT3 decreased (Figure 6B). The cell proliferation ability test results showed that the cell proliferation ability was enhanced after transfection of SNHG16 overexpression plasmid. However, the proliferation ability of cells co-transfected with SNHG16 overexpressing plasmid and sh-STAT3 was decreased (Figure 6C,6D). The changes of cell invasion and migration ability showed that the invasion and migration ability of cells were enhanced after transfection of SNHG16 overexpressed plasmid. However, the invasion and migration abilities of cells co-transfected with SNHG16 overexpressing plasmid and sh-STAT3 were decreased (Figure 6E,6F).
Discussion
During peripheral nerve injury and regeneration, SC function becomes very active (20). SCs are the only cells that divide and proliferate in Wallerian degeneration, are the main cellular source of some active substances (21), and are involved in the whole process of injury and regeneration at the beginning of the Wallerian denaturing response. SCs proliferate and divide immediately after peripheral nerve injury (22), and the regeneration of injured nerves depends largely on their large numbers to form a basal membrane which can prevent adhesion between injured nerves and surrounding tissues. The biological activity of SCs can be enhanced by various methods and approaches to promote the repair of peripheral nerves, which can provide a new theoretical basis and treatment methods for these injuries (23).
The proliferation and migration of SCs contribute to the growth and functional recovery of axons after peripheral nerve injury (24), and studies have shown that lncRNAs can act on SCs to promote their regeneration (25). The expression of Nuclear-enriched Transcript1 (NEAT1) and Special AT-Rich Sequence Binding Protein 1 (SATB1) of lncRNAs increased and the expression of miR-34a decreased after peripheral nerve injury. Overexpression of NEAT1 also promoted the proliferation and migration of SCs. As a competitive endogenous RNA, NEAT1 regulates the expression of nuclear matrix binding region protein 1 through sponge-like interaction with miR-34a. NEAT1 promotes axonal growth of dorsal root ganglion neurons by regulating the expression of miR-34a and nuclear matrix binding region protein 1 (26).
In this study, we found the expression level of lncRNA SNHG16 decreased after sciatic nerve injury and the up-regulation of SNHG16 could promote SC migration and affect STAT3 expression by acting on miR-93-5p. Cell experiments showed that overexpression of SNHG16 promoted the proliferation, migration and invasion of SCs in the sciatic nerve injury model and interaction between SNHG16 and miR-93-5p was predicted by online software analysis. Further experiments confirmed that the expression of miR-93-5p was up-regulated in nerve injury. We confirmed SNHG16 inhibits the effect of miR-93-5p on downstream target genes by adsorption with miR-93-5p, and MiR-93-5p affects the proliferation and migration of SCs by targeting STAT3. Further, we showed that miR-93-5p was highly expressed in nerve injury and was negatively correlated with SNHG16. STAT3 is the target gene of miR-93-5p, and SNHG16/miR-93-5p/STAT3 constitutes the regulatory mechanism of ceRNA. Knocking down STAT3 significantly inhibited the role of SNHG16 in SCs. Many miRNAs also inhibit SC proliferation and migration, and MiR-182 directly targets fibroblast growth factor 9. The inhibitory effect of miR-182 on fibroblast growth factor 9 directly inhibited the migration and proliferation of SCs (27). MiR-9 inhibited the migration of normal and tumor cells, and its silencing promoted SC migration after peripheral nerve injury. This is because collagen triple helix repeat protein 1 is a direct target of miR-9, which inactivates RAC1GTP (28).
Signal activator of transcription (STAT) is an intracellular signal transduction pathway present in the axons and cell bodies of normal peripheral nerves. It is also closely related to the degeneration and regeneration of neurons after peripheral nerve injury (29,30). STAT3, a member of the STAT family, is a key molecule of signal transduction in ciliary neurotrophic factor (CNTF) cells, and in this study, we found that SNHG16 promoted the proliferation, invasion, and migration of SCs by upregulating STAT3. Our results suggest that STAT3 activation is closely related to axon regeneration and SC proliferation and migration in damaged peripheral nerves, especially in migrating SCs.
Conclusions
In summary, lncRNA SNHG16 was studied by using a rat sciatic nerve injury model. We confirmed the down-regulation of lncRNA SNHG16 after peripheral nerve injury, and further experiments showed that silencing lncRNA SNHG16 in SC cells promoted their proliferation, invasion, and migration. Studies on this mechanism suggest that lncRNA SNHG16 may play this role by competing with miR-93-5p to regulate STAT3 expression in SCs. This study provides a new mechanism and a potential therapeutic target for peripheral nerve injury.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-3971
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-3971
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-3971). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license (No.: 2020-012) granted by ethics board of the Second Affiliated Hospital of Soochow University, in compliance with USA Institute for Laboratory Animal Research (ILAR) Guide for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Vijayavenkataraman S. Nerve guide conduits for peripheral nerve injury repair: A review on design, materials and fabrication methods. Acta Biomater 2020;106:54-69. [Crossref] [PubMed]
- Hewson DW, Bedforth NM, Hardman JG. Peripheral nerve injury arising in anaesthesia practice. Anaesthesia 2018;73:51-60. [Crossref] [PubMed]
- Kubiak CA, Kung TA, Brown DL, et al. State-of-the-Art Techniques in Treating Peripheral Nerve Injury. Plast Reconstr Surg 2018;141:702-10. [Crossref] [PubMed]
- Barton MJ, John JS, Clarke M, et al. The Glia Response after Peripheral Nerve Injury: A Comparison between Schwann Cells and Olfactory Ensheathing Cells and Their Uses for Neural Regenerative Therapies. Int J Mol Sci 2017;18:287. [Crossref] [PubMed]
- Liu CY, Yin G, Sun YD, et al. Effect of exosomes from adipose-derived stem cells on the apoptosis of Schwann cells in peripheral nerve injury. CNS Neurosci Ther 2020;26:189-96. [Crossref] [PubMed]
- Yao C, Wang Y, Zhang H, et al. lncRNA TNXA-PS1 Modulates Schwann Cells by Functioning As a Competing Endogenous RNA Following Nerve Injury. J Neurosci 2018;38:6574-85. [Crossref] [PubMed]
- Wang H, Wu J, Zhang X, et al. Microarray analysis of the expression profile of lncRNAs reveals the key role of lncRNA BC088327 as an agonist to heregulin-1β-induced cell proliferation in peripheral nerve injury. Int J Mol Med 2018;41:3477-84. [Crossref] [PubMed]
- Pan B, Zhou HX, Liu Y, et al. Time-dependent differential expression of long non-coding RNAs following peripheral nerve injury. Int J Mol Med 2017;39:1381-92. [Crossref] [PubMed]
- Yao C, Chen Y, Wang J, et al. LncRNA BC088259 promotes Schwann cell migration through Vimentin following peripheral nerve injury. Glia 2020;68:670-9. [Crossref] [PubMed]
- Wu S, Bono J, Tao YX. Long noncoding RNA (lncRNA): a target in neuropathic pain. Expert Opin Ther Targets 2019;23:15-20. [Crossref] [PubMed]
- Yao C, Wang J, Zhang H, et al. Long non-coding RNA uc.217 regulates neurite outgrowth in dorsal root ganglion neurons following peripheral nerve injury. Eur J Neurosci 2015;42:1718-25. [Crossref] [PubMed]
- Costigan M, Belfer I, Griffin RS, et al. Multiple chronic pain states are associated with a common amino acid-changing allele in KCNS1. Brain 2010;133:2519-27. [Crossref] [PubMed]
- Cai C, Huo Q, Wang X, et al. SNHG16 contributes to breast cancer cell migration by competitively binding miR-98 with E2F5. Biochem Biophys Res Commun 2017;485:272-8. [Crossref] [PubMed]
- Cao X, Xu J, Yue D. LncRNA-SNHG16 predicts poor prognosis and promotes tumor proliferation through epigenetically silencing p21 in bladder cancer. Cancer Gene Ther 2018;25:10-7. [Crossref] [PubMed]
- Scheuermann JC, Boyer LA. Getting to the heart of the matter: long non-coding RNAs in cardiac development and disease. EMBO J 2013;32:1805-16. [Crossref] [PubMed]
- Yu M, Ohira M, Li Y, et al. High expression of ncRAN, a novel non-coding RNA mapped to chromosome 17q25.1, is associated with poor prognosis in neuroblastoma. Int J Oncol 2009;34:931-8. [PubMed]
- Christensen LL, True K, Hamilton MP, et al. SNHG16 is regulated by the Wnt pathway in colorectal cancer and affects genes involved in lipid metabolism. Mol Oncol 2016;10:1266-82. [Crossref] [PubMed]
- Tao Y. Isolation and culture of Schwann cells. Neural Development Springer, 2013:93-104.
- Jin YQ, Liu W, Hong TH, et al. Efficient Schwann cell purification by differential cell detachment using multiplex collagenase treatment. J Neurosci Methods 2008;170:140-8. [Crossref] [PubMed]
- Han GH, Peng J, Liu P, et al. Therapeutic strategies for peripheral nerve injury: decellularized nerve conduits and Schwann cell transplantation. Neural Regen Res 2019;14:1343-51. [Crossref] [PubMed]
- Nocera G, Jacob C. Mechanisms of Schwann cell plasticity involved in peripheral nerve repair after injury. Cell Mol Life Sci 2020;77:3977-89. [Crossref] [PubMed]
- Pan B, Shi ZJ, Yan JY, et al. Long non-coding RNA NONMMUG014387 promotes Schwann cell proliferation after peripheral nerve injury. Neural Regen Res 2017;12:2084-91. [Crossref] [PubMed]
- Yi S, Liu Q, Wang X, et al. Tau modulates Schwann cell proliferation, migration and differentiation following peripheral nerve injury. J Cell Sci 2019;132:jcs222059 [Crossref] [PubMed]
- Wang Y, Shan Q, Pan J, et al. Actin Cytoskeleton Affects Schwann Cell Migration and Peripheral Nerve Regeneration. Front Physiol 2018;9:23. [Crossref] [PubMed]
- Yu B, Zhou S, Yi S, et al. The regulatory roles of non-coding RNAs in nerve injury and regeneration. Prog Neurobiol 2015;134:122-39. [Crossref] [PubMed]
- Liu X, Yu X, He Y, et al. Long noncoding RNA nuclear enriched abundant transcript 1 promotes the proliferation and migration of Schwann cells by regulating the miR-34a/Satb1 axis. J Cell Physiol 2019; Epub ahead of print. [Crossref] [PubMed]
- Kouri FM, Hurley LA, Daniel WL, et al. miR-182 integrates apoptosis, growth, and differentiation programs in glioblastoma. Genes Dev 2015;29:732-45. [Crossref] [PubMed]
- Zhou S, Gao R, Hu W, et al. MiR-9 inhibits Schwann cell migration by targeting Cthrc1 following sciatic nerve injury. J Cell Sci 2014;127:967-76. [Crossref] [PubMed]
- Yao GL, Kato H, Khalil M, et al. Selective upregulation of cytokine receptor subchain and their intracellular signalling molecules after peripheral nerve injury. Eur J Neurosci 1997;9:1047-54. [Crossref] [PubMed]
- Rajan P, Stewart CL, Fink JS. LIF-mediated activation of STAT proteins after neuronal injury in vivo. Neuroreport 1995;6:2240-4. [Crossref] [PubMed]
(English Language Editor: B. Draper)


