
The methylation modification of m6A regulators contributes to the prognosis of head and neck squamous cell carcinoma
Introduction
Head and neck cancer, ranked as the 7th most common cancer worldwide in 2018, accounted for 3% of all cancer cases (1). Head and neck squamous cell carcinoma (HNSCC) refers to squamous cell carcinoma arising from mucosal surfaces of the sinonasal cavity, oral cavity, larynx, and pharynx, but does not include nasopharyngeal cancer (2). Improvements in surgery and radiotherapy techniques and curative multidisciplinary therapies have substantially enhanced patient outcomes (3). However, recurrent or metastatic disease occurred in almost more than 65% HNSCC patients results in dismal prognosis (4). Thus, the introduction of new treatment options targeted at vital new regulators of carcinogenesis is urgent.
N6-methyladenosine (m6A) modification, which is defined as the addition of a methyl group at the N6 site of adenosine, is the most prevalent internal chemical modification in eukaryotes (5). Consisting of a group of proteins referred to as the “writer”, “eraser”, and “reader”, m6A methylation can affect multiple aspects of ribonucleic acid (RNA) metabolism, including messenger RNA (mRNA) splicing, stability, localization, and translation (6). The methylation of m6A mRNA is mainly accomplished by “writer” proteins, including methyltransferase-like 14 (METTL14), Wilms’ tumor 1-associating protein (WTAP) and methyltransferase-like 3 (METTL3) (7). Recently, RNA-binding motif protein 15 (RBM15), KIAA1429, and zinc finger CCCH domain-containing protein 13 (ZC3H13) were added to the methyltransferase complex (8). Conversely, the demethylation procedure is mainly conducted by “eraser” proteins, including AlkB homolog 1 (ALKBH1), AlkB homolog 5 (ALKBH5), and the fat mass and obesity-associated protein (FTO) (9,10). More importantly, the exertion of m6A modification in the degradation and translation of downstream RNA mainly relies on the recruitment of “reader” proteins, including five YT521-B homology (YTH) domain family members (YTHDF1–3 and YTHDC1–2), and heterogeneous nuclear ribonucleoproteins (11,12). The “writer”, “eraser”, and “reader” genes collaboratively control reversible m6A modification, and thus play vital roles in physiological activities and human diseases, especially cancer (13).
More and more evidence suggest that m6A RNA methylation regulators play a vital role in cancer development and prognosis prediction; however, little is known about the relationship between m6A modification and HNSCC. Two previous studies have developed two different two-gene panel to predict the patient outcome in HNSCC. However, the relationship between m6A RNA methylation modulators signature and prognosis of HNSCC still needs further verification. To further investigate the precise m6A regulation pattern in HNSCC, we systematically analyzed the expression of those well studied m6A regulators in HNSCC and studied the correlation between these regulators and the clinicopathological features of patients using patient data from The Cancer Genome Atlas (TCGA) database. More importantly, we constructed a risk signature comprising three regulators rather than different two gene panel to classify the prognosis of HNSCC patients. We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/atm-21-4077).
Methods
Data collection
The RNA-seq transcriptome data of 422 HNSCC patients and their corresponding clinical and prognosis information were acquired from TCGA database (https://cancergenome.nih.gov/). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
m6A RNA methylation regulators selection
Thirteen genes are recognized as vital m6A methylation regulators, including METTL3, METTL14, WTAP, ALKBH5, FTO, YTHDC1, YTHDC2, YTHDF1, YTHDF2, HNRNPC, KIAA1429, RBM15, and ZC3H13. However, due to the KIAA1429 data missing, we could only investigate the relationship among the other 12 m6A-related genes and the clinicopathological characteristics as well as the overall survival (OS) of HNSCC patients based on TCGA dataset.
Bioinformatics analysis
The relationship between the expressions of m6A RNA methylation regulators and clinicopathological variables in HNSCC was analyzed by the Limma package (http://www.bioconductor.org/packages/release/bioc/html/limma.html) with a cut-off P value of 0.05. Next, vioplot was used to visualize the expression of the 12 regulators in 381 tumor tissues and 41 normal tissues. A spearman analysis was conducted to explore the correlations among these regulator genes. Next, the Consensus Cluster Plus package was used (https://www.bioconductor.org/packages/release/bioc/html/ConsensusClusterPlus.html) to divide tumor samples into two groups, and a principal component analysis (PCA) was conducted to verify the grouping results. The analysis of the survival of the two clusters was processed by a survival package. To explore the prognostic role of m6A methylation regulators in HNSCC patients, we performed a univariate Cox analysis and developed a risk signature by employing the least absolute shrinkage and selection operator (LASSO) Cox regression algorithm. Three genes were identified as powerful prognostic factors. The risk score of each patient was calculated using the following formula:
Coefi is the coefficient value and xi is the expression value of each selected gene.
Statistical analysis
The Wilcoxon’s test was used to compare the expression level of the 12 m6A RNA methylation regulators between the tumor and normal tissues. A one-way analysis of variance was used to analyze the relationship between the m6A regulators and the clinicopathological features of HNSCC patients. The median risk score was set as the cut-off value to divide patients into the high- or low-risk group. To further analyze the OS difference between the two groups, the Kaplan-Meier method was used. R software (version 3.5.1) was used for all the statistical analyses. P values of less than 0.05 were considered statistically significant.
Results
The expression landscape of m6A RNA methylation regulators in HNSCC
To better understand the role of m6A RNA methylation regulators in carcinogenesis, we first compared the m6A RNA methylation regulators expression between cancer tissues and normal tissues based on the extracted RNA data from TCGA database, and found eight differentially expressed regulators. The heatmap and the vioplot both showed that FTO (P<0.01), ALKBH5 (P<0.01), YTHDF1 (P<0.001), METTL3 (P<0.001), HNRNPC (P<0.001), WTAP (P<0.001), and RBM15 (P<0.001) were significantly upregulated in cancer tissues (see Figure 1A,1B). Additionally, YTHDC2 expression was significantly lower in cancer tissues than normal tissues (P<0.01; see Figure 1A,1B). The analysis of these m6A-related genes revealed that the strongest correlations were between METTL14 and YTHDC1, and METTL14 and YTHDF2 (see Figure 1C).
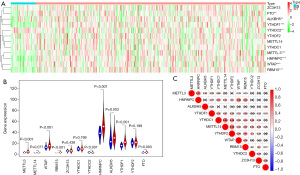
Relationship between the expression of the m6A RNA methylation regulators and the clinicopathological features of HNSCC patients
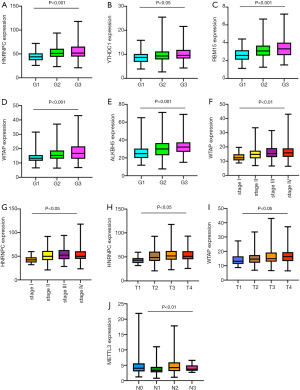
To examine the relationship between the m6A RNA methylation regulators and the clinicopathological features of HNSCC patients, we analyzed the clinical significance of these regulators individually. The results showed that the expression of HNRNPC (P<0.001), YTHDC1 (P<0.05), RBM15 (P<0.001), and WTAP (P<0.001) was significantly correlated with grade (see Figure 2A-2E). Additionally, the expression of HNRNPC, YTHDC1, RBM15, and WTAP were elevated as the grade increased. Further, the expression of WTAP and HNRNPC were also significantly correlated with grade and tumor (T) stage (see Figure 2F-2I). However, METTL3 expression was negatively correlated with nodes (N) stage (P<0.01; see Figure 2J).
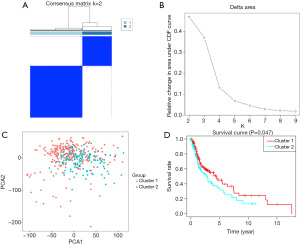
Cluster classification based on m6A RNA methylation expression
The Consensus Cluster Plus package was used to group the 381 HNSCC cancer tissues. Based on the cumulative distribution function value, we tried to divide these samples into two or three groups. We found that dividing these samples into two groups ensured the significant difference between groups (see Figure 3A,3B). We then used PCA to verify the classification. The results showed that Clusters 1 and 2 gathered together respectively (see Figure 3C). To further understand the relationship between clustering and clinical outcomes, we analyzed the OS data of these two clusters. We found that the Cluster 1 subgroup had a higher OS than the Cluster 2 subgroup (see Figure 3D).
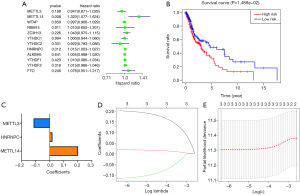
Prognostic role of m6A RNA methylation regulators in HNSCC
Next, we conducted a Cox univariate analysis to explore the prognostic role of m6A RNA methylation regulators in HNSCC (see Figure 4A). Notably, the high expression of METTL14 resulted in worse survival in HNSCC patients [hazard ratio (HR) =1.323, 95% confidence interval (CI) =1.077–1.624]. Among these m6A regulators, METTL3, METTL14, and HNRNPC were selected to build the risk signature to predict prognosis according to the P value from the previous univariate analysis. To verify the risk signature prediction role, the cancer patients were divided into high- and low-risk groups based on the median score. The OS curve indicated that the high-risk group had a worse survival rate than the low-risk group (see Figure 4B). We next used the coefficients of the three regulators obtained from the LASSO regression algorithm to calculate the risk scores of HNSCC patients from TCGA dataset (see Figure 4C-4E).
The risk score was closely related to the clinical outcomes of HNSCC patients
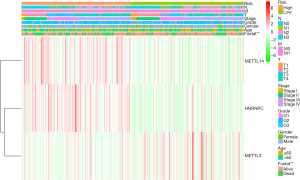
We also generated a heat map to investigate the correlation between the risk score and clinicopathological features of HNSCC patients from TCGA datasets. We only found a significant difference between the high- and low-risk group in living status (P<0.01; see Figure 5). Further, the high-risk group had a higher proportion of METTL14 and HNRNPC and a lower proportion of METTL3 than the low-risk group (see Figure 5).
Discussion
There is evidence that up to 60% of patients with HNSCC are diagnosed at an advanced stage despite improvements in screening and epidemiology changes. Advanced stage HNSCC is characterized by local invasion, metastases to the regional nodes or even distant metastasis. Thus, advanced stage HNSCC carries a high local recurrence rate and poor prognosis, especially in patients with laryngeal and hypopharyngeal cancer (14). Patients in the same late stage could have different reactions to the same treatment strategy, which could result in distinct outcomes (15). In this case, the tumor, nodes, metastases (TNM) stage alone could not sufficiently predict patient outcomes, or direct personalized targeted therapy. Thus, new gene regulators in tumorigenesis urgently need to be identified to group patients and ensure the optimal selection of treatments.
In recent years, the role of m6A modification in many biological processes such as immune regulation, metabolism, maintenance and differentiation of cell dryness has been proved. In addition, numerous studies have found that m6A modification of RNA also plays an important regulatory role in cancer development by regulating oncoprotein expression, intriguing cell proliferation and tumor progression (16,17). In HNSCC, the regulatory role of m6A modification in cancer pathogenesis has also been proved. METTL3 and METTL 14 mediated m6A modification can stabilize lncAROD, which protected YBX1 from proteasomal degradation in HNSCC, thus helping lncAROD to exert its oncogenic role (18). What’s more, METTL3 can promote EZH2 expression and NPC progression (19). METTL3 can interact with IGF2BP1, which promotes BMI-1 expression and accelerates OSCC proliferation and metastasis (20). Since HNSCC contains multiple cancer sites and some genes showed contradictory roles in different cancer type, bioinformatics analysis turns to be an effective way to explore the core genes of HNSCC and provide potential target for tumor treatment. In our study, we used TCGA dataset to extract patients’ complete data with detailed information. What’s more, we went through all data carefully and delete some patients for important data missing to ensure the fidelity of our bioinformatics analysis. We developed a three gene panel as the prognostic characteristic based on their relationship with tumor grade, clinical stage, T stage, N stage and OS, thus offered three important candidate gene for further investigation.
We first analyzed the different expression pattern between cancer tissues and normal tissues and the relationship among the regulators. The results showed that eight of the 12 regulators (i.e., ALKBH5, FTO, YTHDF1, YTHDC2, METTL3, HNRNPC, WTAP, and RBM15) were significantly differently expressed in cancer tissues compared to normal tissues. With the exception of YTHDC2, all the other seven regulators exhibited higher expression levels in cancer tissues. However, a TCGA data analysis of 508 HNSCC cancer patients, indicated that the YTHDC2 expression in tumors was higher compared to normal ones. In addition, relatively high YTHDC2 expression was correlated with poorer survival (21). YTHDC2 has been recognized as a frequently altered regulator in different cancer types, and is mainly involved in carcinogenesis by increasing hypoxia-inducible factor-1α translation (22). These controversial results may be attributable to the heterogeneity of cancers arising from different sites.
We also found that some of the m6A RNA methylation regulators were significantly associated with different clinicopathological features in HNSCC. HNRNPC is a RNA-binding protein responsible for pre-mRNA processing that has been reported to promote chemoresistance in gastric cancer and facilitate colorectal cancer progression (23,24). Our results showed that HNRNPC was not only highly expressed in cancer tissues, but was also significantly associated with tumor grade, tumor clinical stage, and T stage. Consistent with HNRNPC oncogenic function, higher expression of HNRNPC showed in late stage. WTAP is recognized as a m6A writer that contributes to the METTL3-METTL14 methyltransferase localization to the nuclear and executes translation and post-translation regulations (25,26). It can facilitate metastasis in pancreatic cancer and promote the progression of hepatocellular carcinoma (HCC) (27,28). In renal cell carcinoma, WTAP has also been shown to play an oncogenic role (29). In HNSCC, we observed that WTAP expression was correlated with tumor grade, tumor clinical stage, and tumor T stage, and mainly functioned as a tumor promoter.
To examine the prognostic value of the m6A RNA methylation regulators, a Cox univariate analysis and LASSO regression analysis were undertaken and three regulators were selected to construct a risk signature. Next, we classified the patients into high- or low-risk groups according to their risk scores. The OS curve verified that the risk signature could help distinguish the patients’ outcomes. The heatmap showed that METTL14 tended to have a higher level of expression in the high-risk group than the low-risk group. However, METTL3 in the high-risk group exhibited a lower level expression pattern. METTL3 is the main component of the “writer” complex that has been proven to be closely related to cancer progression. METTL3 has been reported to exhibit significantly higher levels of expression in HCC compared to normal tissues. Moreover, it is critical to epithelial mesenchymal transition in HCC (30). In gastric cancer, METTL3 has also been shown to have a higher expression pattern in cancer tissues than normal tissues, and to be a poor prognostic indicator in gastric cancer patients (31). Conversely, of all the m6A gene regulators we analyzed, METTL3 was the sole 1 associated with lymph node stage, and was also negatively correlated to N stage, which is inconsistent with the oncogene role it has been verified to have in other cancer types. The favorable prognostic role of METTL3 in HNSCC needs to be verified in cancer tissues and other public databases.
More importantly, the landmark discovery of the role of immune check point in immune evasion of cancer cells brought immunotherapy era to the cancer treatment. The OS benefit from immunotherapy (i.e., pembrolizumab, nivolumab, camrelizumab) has been proved in multiple phase II to III clinical trial especially in recurrent or metastatic HNSCC. To better understand the tumor response to immune therapy, tumor microenvironment is the key part to draw the whole immune map. Tumor microenvironment is formed from dynamic changes involving multiple immunosuppressive signal pathways and values a lot in predicting patient prognosis and therapeutic response (32). Previous study indicated that lower level of METTL3 and METTL14 contributes to the T cells differentiation (33). What’s more, CD8+ T cells and NK cells showed increased level in YTHDF1-deficient mouse tumors (34). In HNSCC, tumor infiltrating lymphocytes related immune score has been proved to potentially predict the treatment efficacy. Yi et al. demonstrated that a risk score consisted of seven m6A genes was positively correlated with dendritic cells, neutrophils infiltration and negatively correlated with CD4+ T, CD8+ T and B cells infiltration. They also suggested that up-regulated m6A modulators was positively correlated with PD-L1 in HNSCC tumor immune microenvironment, which indicated m6A modulators as potential immunotherapy target in HNSCC (35). Our study provided three potential target genes for further immunotherapy investigation for HNSCC treatment.
Conclusions
Our study showed that there was a close relationship between the expression of m6A RNA methylation regulators and the clinicopathological features of HNSCC patients. Additionally, we constructed a risk signature comprising three regulators that could serve as the prognostic predictor in HNSCC. Our results provide vital evidence that can be used to further predict cancer prognosis and tumor response to cancer treatment of HNSCC.
Acknowledgments
Funding: This study was funded by the Natural Science Foundation of Zhejiang Province (grant number: LQ19H160033).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-4077
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-4077). Dr. YC reported that this study was funded by the Natural Science Foundation of Zhejiang Province (grant number: LQ19H160033). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The Research Ethics Committee of The First Affiliated Hospital of Zhejiang University School of Medicine confirmed that no ethical approval was required and waived informed consent. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Chow LQM. Head and neck cancer. N Engl J Med 2020;382:60-72. [Crossref] [PubMed]
- Eskander A, Irish J, Groome PA, et al. Volume-outcome relationships for head and neck cancer surgery in a universal health care system. Laryngoscope 2014;124:2081-8. [Crossref] [PubMed]
- Argiris A, Karamouzis MV, Raben D, et al. Head and neck cancer. Lancet 2008;371:1695-709. [Crossref] [PubMed]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012;485:201-6. [Crossref] [PubMed]
- Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014;505:117-20. [Crossref] [PubMed]
- Wang X, Feng J, Xue Y, et al. Corrigendum: structural basis of N6-adenosine methylation by the METTL3-METTL14 complex. Nature 2017;542:260. [Crossref] [PubMed]
- Wen J, Lv R, Ma H, et al. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol Cell 2018;69:1028-38.e6. [Crossref] [PubMed]
- Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 2013;49:18-29. [Crossref] [PubMed]
- Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 2011;7:885-7. [Crossref] [PubMed]
- Shi H, Wang X, Lu Z, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res 2017;27:315-28. [Crossref] [PubMed]
- Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 2012;44:694-8. [Crossref] [PubMed]
- Chen J, Du B. Novel positioning from obesity to cancer: FTO, an m6A RNA demethylase, regulates tumour progression. J Cancer Res Clin Oncol 2019;145:19-29. [Crossref] [PubMed]
- Braakhuis BJ, Brakenhoff RH, Leemans CR. Treatment choice for locally advanced head and neck cancers on the basis of risk factors: biological risk factors. Ann Oncol 2012;23:x173-7. [Crossref] [PubMed]
- Brana I, Siu LL. Locally advanced head and neck squamous cell cancer: treatment choice based on risk factors and optimizing drug prescription. Ann Oncol 2012;23:x178-85. [Crossref] [PubMed]
- Zhou S, Bai ZL, Xia D, et al. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β-catenin through mRNA demethylation. Mol Carcinog 2018;57:590-7. [Crossref] [PubMed]
- Ge L, Zhang N, Chen Z, et al. Level of N6-methyladenosine in peripheral blood RNA: a novel predictive biomarker for gastric cancer. Clin Chem 2020;66:342-51. [Crossref] [PubMed]
- Ban Y, Tan P, Cai J, et al. LNCAROD is stabilized by m6A methylation and promotes cancer progression via forming a ternary complex with HSPA1A and YBX1 in head and neck squamous cell carcinoma. Mol Oncol 2020;14:1282-96. [Crossref] [PubMed]
- Meng QZ, Cong CH, Li XJ, et al. METTL3 promotes the progression of nasopharyngeal carcinoma through mediating M6A modification of EZH2. Eur Rev Med Pharmacol Sci 2020;24:4328-36. [PubMed]
- Liu L, Wu Y, Li Q, et al. METTL3 promotes tumorigenesis and metastasis through BMI1 m6A methylation in oral squamous cell carcinoma. Mol Ther 2020;28:2177-90. [Crossref] [PubMed]
- Zhou X, Han J, Zhen X, et al. Analysis of genetic alteration signatures and prognostic values of m6A regulatory genes in head and neck squamous cell carcinoma. Front Oncol 2020;10:718. [Crossref] [PubMed]
- Tanabe A, Tanikawa K, Tsunetomi M, et al. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1α mRNA is translated. Cancer Lett 2016;376:34-42. [Crossref] [PubMed]
- Huang H, Han Y, Zhang C, et al. HNRNPC as a candidate biomarker for chemoresistance in gastric cancer. Tumour Biol 2016;37:3527-34. [Crossref] [PubMed]
- Liu X, Liu L, Dong Z, et al. Expression patterns and prognostic value of m6A-related genes in colorectal cancer. Am J Transl Res 2019;11:3972-91. [PubMed]
- Horiuchi K, Umetani M, Minami T, et al. Wilms' tumor 1-associating protein regulates G2/M transition through stabilization of cyclin A2 mRNA. Proc Natl Acad Sci U S A 2006;103:17278-83. [Crossref] [PubMed]
- Ping XL, Sun BF, Wang L, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 2014;24:177-89. [Crossref] [PubMed]
- Chen Y, Peng C, Chen J, et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol Cancer 2019;18:127. [Crossref] [PubMed]
- Li BQ, Liang ZY, Seery S, et al. WT1 associated protein promotes metastasis and chemo-resistance to gemcitabine by stabilizing Fak mRNA in pancreatic cancer. Cancer Lett 2019;451:48-57. [Crossref] [PubMed]
- Tang J, Wang F, Cheng G, et al. Wilms' tumor 1-associating protein promotes renal cell carcinoma proliferation by regulating CDK2 mRNA stability. J Exp Clin Cancer Res 2018;37:40. [Crossref] [PubMed]
- Chen M, Wei L, Law CT, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 2018;67:2254-70. [Crossref] [PubMed]
- Liu T, Yang S, Sui J, et al. Dysregulated N6-methyladenosine methylation writer METTL3 contributes to the proliferation and migration of gastric cancer. J Cell Physiol 2020;235:548-62. [Crossref] [PubMed]
- Fridman WH, Pagès F, Sautès-Fridman C, et al. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012;12:298-306. [Crossref] [PubMed]
- Li HB, Tong J, Zhu S, et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 2017;548:338-42. [Crossref] [PubMed]
- Han D, Liu J, Chen C, et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature 2019;566:270-4. Erratum in: Nature 2019;568:E3. [Crossref] [PubMed]
- Yi L, Wu G, Guo L, et al. Comprehensive analysis of the PD-L1 and immune infiltrates of m6A RNA methylation regulators in head and neck squamous cell carcinoma. Mol Ther Nucleic Acids 2020;21:299-314. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


