
This article has an erratum available at: http://dx.doi.org/10.21037/atm-2023-13 the article has been update on 2023-06-16 at here.
Involvement of ferroptosis in the granulosa cells proliferation of PCOS through the circRHBG/miR-515/SLC7A11 axis
Introduction
Polycystic ovary syndrome (PCOS) is a heterogeneous disease, which is defined as excessive androgen, ovarian dysfunction while other specific diagnoses need to be ruled out (1). There's no test to definitively diagnose PCOS, three PCOS diagnostic standards currently exist (1-3). The incidence of PCOS in premenopausal women is reported to be 6–20%, and it is the most common endocrine disease in adult women (4,5). The cause of PCOS has not yet been clarified to a large extent, and increasing evidence shows that PCOS is a complex polygenic genetic disease that is greatly affected by environmental factors (6,7). PCOS is related to abdominal obesity, insulin resistance, obesity, metabolic disorders, and cardiovascular risk factors (3,8). With the development of next-generation sequencing technology, more and more genes involved in regulating the progression of PCOS have been discovered (9,10).
Circular RNA (circRNA) can be synthesized by reverse splicing of coding gene exons or classical mRNA splicing without degradation of the intron lasso (11,12). In addition to the effect of exon reverse splicing on its parental gene mRNA splicing, studies have reported that circRNA can regulate gene expression in the nucleus, competitively binding miRNA or protein, and acts as the backbone of the circRNA-protein complex (13-16). Many studies have confirmed the role of circRNA in various diseases; for example, circRNA HIPK3 causes endothelial proliferation and vascular dysfunction in diabetic retinopathy (17). Cancer-specific PML-RARA translocation-derived fusion circRNA can also promote transformation and cell survival (18). The role and mechanism of circRNA in PCOS has gradually become a research hotspot (17,19).
As a common type of non-coding RNA, many microRNAs (miRNAs) have also been found to play a role in the occurrence and development of PCOS. Studies have found that, compared with non-PCOS women, the expression of miRNA in the follicular fluid of PCOS women is very different, and the expression of potential target genes related to the PCOS phenotype has also changed (20,21). Some researchers have constructed a circRNA-microRNA interaction network. The results showed that the differentially expressed circRNA in PCOS is related to inflammation, bacterial infection, and oxidative stress (22). However, to date, these studies have only investigated discrepancy of circRNAs between PCOS patients and controls but have not further studied the specific mechanism. The circRHBG-microRNA pathway in PCOS has not been well studied.
In the present study, we screened out circRHBG with elevated expression in PCOS patients through circRNA microarray and speculated that it plays an important role in PCOS. Bioinformatics analysis was then applied to predict the downstream target of circRHBG, and we found that miR-515 is a potential target of it. In addition, we found that the upregulation of circRHBG expression inhibited ferroptosis in KGN and SVOG cells through the circRHBG/miR-515/SLC7A11 axis. Our results partially proved the role of the circRHBG/miR-515/SLC7A11 axis in PCOS, which may help in the clinical diagnosis and treatment of PCOS. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/atm-21-4174).
Methods
Clinical sample data and ethics approval
We recruited 12 infertility patients (6 PCOS and 6 controls) aged 20 and 38 who underwent in vitro fertilization (IVF) at the Department of Reproductive Medicine Center, the First Affiliated Hospital of Sun Yat-Sen University. We adopted the Rotterdam definition for PCOS, which involves diagnosis with 2 of 3 clinical manifestations, including clinical and/or biochemical hyperandrogenism, ovulation dysfunction, and PCOM after excluding patients with Cushing's syndrome, congenital adrenal hyperplasia, and androgen secreting tumors. Six women without PCOS at the similar age as that of PCOS patients were gathered as the normal control group. The patients with body mass index (BMI) >28 kg/m2, basal FSH level >10 mIU/mL, endometriosis, diminished ovarian reserve, chromosome abnormality, hydrosalpinx, systemic illness or endocrine disorders were excluded from this study. All patients provided signed written informed consent, and all experiments were performed under the approval of the local ethics committee of the First Affiliated Hospital of Sun Yat-Sen University ([2021]399-1). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
All patients were treated the antagonist stimulation protocol. Briefly, on cycle day 2, ovarian stimulation was started by daily injection of recombinant FSH (r-FSH) (Gonal-F; Merck Serono). Gonadotropin-releasing hormone (GnRH) antagonist (Cetrotide, 0.25 mg; Merck Serono) injection was started from the day 6 of stimulation. When at least three follicles had reached 17 mm or two follicles had reached 18 mm in diameter, an intramuscular injection of 5,000 IU-10000 IU of human chorionic gonadotropin (hCG) is used for triggering final oocyte maturation. Oocyte retrieval was scheduled approximately 34–36 h after hCG injection by transvaginal ultrasound-guided follicular aspiration. Follicle aspiration of mature oocytes was used to obtain ovarian granulosa cells from all 6 samples. Clear follicular fluid containing granulosa cells was collected and centrifuged at 626 ×g for 10 min. The cells were resuspended in hyaluronidase and digested at 37 °C for 20 min. Finally, the lymphocyte separation solution was added and centrifuged at 626 ×g for 10 min. The white turbid clusters of cells in the middle were granular cells.
circRNA microarray
CapitalBio Technology Human CircRNA Array v2 was designed with 4 identical arrays per slide, with each array containing probes analyzing about 170,340 human circRNAs.
Cell culture and transfection
KGN and SVOG cells (GC cell line) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). All cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum (Gibco, Grand Island, NY, USA, Cat.# 16140071) and 1% gentamicin at 37 °C in 5% CO2. Lipofectamine 3000 reagent (Invitrogen, Waltham, USA) was used to transfect all plasmids, siRNA, and negative controls purchased from GeneChem (Shanghai, China), according to the manufacturer's instructions.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis
TRIzol reagent (Life Technologies, Carlsbad, CA, USA) was used to isolate and extract total RNA from cells. SYBR Premix Ex Taq II kit (Takara Bio, Dalian, China, Cat.# RR036A) was then used to synthesize cDNA. SYBR Green PCR Master Mix (Takara Bio, Dalian, China, Cat.# RR420A) was used to amplify the target RNA on the CFX96 Real-time PCR System (Bio-Rad, Hercules, CA, USA), and β-actin was selected as the internal control. The expression change of the target gene was calculated using the 2-ΔΔCt method.
Luciferase reporter assay
The WT and MUT luciferase reporter vectors of circRHBG and SLC7A11 3'-untranslated region (UTR) were constructed. After co-transfection of miR-155-5p mimics or miR-155-5p locked nucleic acid (LNA) and their negative control, the dual-luciferase reporter gene detection kit (Promega, Madison, WI, USA) was used to evaluate the luciferase reporter gene activity.
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 assay was used to assess the proliferation ability of cells. CCK-8 solution (10 µL) was added to each well 48 h after transfection. After incubating for 2 h at 37 °C, a microtiter plate reader (Epoch2; BioTek, Winooski, VT, USA) was used to measure the absorbance at 450 nM.
EdU assay
The Cell-Light EdU DNA Cell Proliferation Kit (RiboBio,Guangzhou, China) was used to test cell proliferation. Two days after transfection of KGN and SVOG cells with si-NC and si-RHBG, EdU was added for 48 h. The cells were then fixed with 4% paraformaldehyde and stained with Apollo dye solution (Beyotime, Shanghai, China). Nucleic acids were stained by DAPI. Fluorescence microscope and Image-Pro Plus software were used for the images and to evaluate cellular EdU incorporation.
Western blot
The total protein extracted from the cells was separated by 10% SDS-PAGE gel and transferred to a PVDF membrane. The protein was then placed in 5% skim milk and blocked at room temperature for 1 h. Anti-SLC7A11 (1:1,000; Abcam, Cambridge, MA, USA) or anti-GAPDH (1:1,000; Abcam, Cambridge, MA, USA) primary antibody was then added and incubated at 4 °C overnight. After washing 3 times with TBST, the protein was incubated with secondary antibody for 2 h at room temperature and quantified by chemiluminescence detection.
Detection of glutathione (GSH)/oxidized glutathione (GSSG) ratio
The GSH/GSSG Quantitative Kit II (Dojindo, Shanghai, China) was used to measure the GSH/GSSG ratio. The sample and standard solution were prepared, and concentration detected, according to the kit instructions. Finally, a microplate reader was used to calculate the relative level.
Statistical analysis
All data were expressed as the mean of 3 experiments (biological replicates) ± standard deviation (SD). Student’s t-test and 1-way analysis of variance were used to compare quantitative data. All statistical analyses were performed by using SPSS version 25.0 (IBM, SPSS, Chicago, IL, USA). A 2-sided P value <0.05 was considered statistically significant.
Results
circRHBG upregulation in PCOS
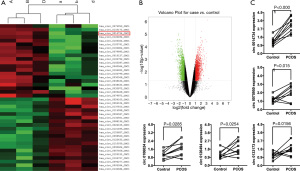
We used circRNA high-throughput chips to screen the differential expression profiles of related circRNA in 6 pairs of granular cells in the PCOS patients and control patients. According to the multiple of the difference greater than 1-fold and the P<0.05 criterion, it was found that there were 575 PCOS-related circRNAs, among which 362 were upregulated circRNA and 213 were downregulated circRNA (Figure 1A,1B). Five upregulated circRNAs were then selected for qRT-PCR verification in 9 pairs of PCOS patients and non-PCOS patients. The results showed that circRNA hsa_circ_0014734 (circRHBG) derived from RHBG was upregulated in 9 cases of granular cells from PCOS patients (9/9: P<0.05), which was significantly higher than that of control group. This was consistent with the microarray results (Figure 1C).
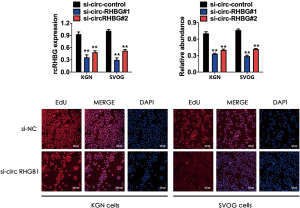
circRHBG knockdown inhibits KGN and SVOG cell proliferation
In order to explore the biological function of circRHBG in PCOS, we used CCK-8 assay and the EdU method to compare the viability of KGN and SVOG cells with different circRHBG expression levels. After transfection of the 2 siRNAs, the circRHBG expression level in KGN and SVOG cells decreased significantly, and the knockdown efficiency of si-circRHGB1 was higher (Figure 2A). The CCK-8 assay results showed that the cell viability of si-circRHBG-transfected cells was significantly lower than that of si-NC transfected cells, and the cell viability of si-circRHGB1-transfected cells was lower than that of si-circRHGB2-transfected cells (Figure 2B). We used si-circRHGB1 with higher knockdown efficiency for the EdU experiments. The EdU test results showed that the EdU incorporation rate of KGN and SVOG cells transfected with si-circRHGB1 was significantly lower than that of cells transfected with si-NC (Figure 2C). Briefly, after knocking down the expression of circRHBG, the proliferation ability of KGN and SVOG cells was significantly weakened.
circRHBG functions as a sponge for miR-515-5p
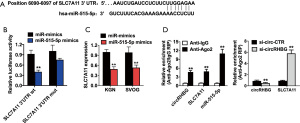
circRNA often regulates gene expression through an endogenous competitive RNA mechanism. In order to explore the downstream target molecules of circRHBG, we used circRNA interactome (https://circinteractome.nia.nih.gov) to predict miRNAs that may bind to it. The results showed that there is a sequence that binds to miR-515-5p on circRHBG (Figure 3A). To further verify the binding of these 2 molecules, we performed the dual-luciferase reporter assay in KGN and SVOG cells. The results showed that the fluorescence intensity of cells transfected with circRHGB-wt + miR-NC mimics was higher than that of cells transfected with circRHGB-wt + miR-515-5p mimics. The fluorescence intensity of cells transfected with circRHGB-mut + miR-NC mimics was not different from that of cells transfected with circRHGB-mut + miR-515-5p mimics (Figure 3B). To further verify the binding of circRHBG to miR-515-5p, we performed MS2bp-MS2bs-based RIP assay in KGN and SVOG cells. The results showed that miR-515-5p was enriched in the MS2bs-circRHBG group (Figure 3C). qRT-PCR showed that, in KGN and SVOG cells, the expression level of miR-515-5p in cells transfected with si-circRHBG was significantly higher than that in cells transfected with si-NC (Figure 3D). These results indicate that circRHBG acts as a miR-515-5p sponge in PCOS.

circRHBG competes with SLC7A11 to bind to miR-515-5p
miRNA usually inhibits the expression of the target gene by binding to the 3'-UTR of the targeted mRNA to degrade it (23). To explore the target molecules of miR-515-5p, we used the TargetScan algorithm to identify SLC7A11 as a candidate target oncogene (Figure 4A). To further verify the combination of these 2 molecules, we used the dual-luciferase reporter assay in KGN cells. The results showed that the fluorescence intensity of cells transfected with SLC7A11-wt + miR-NC mimic was higher than that of cells transfected with SLC7A11-wt + miR-515-5p mimics. The fluorescence intensity of cells transfected with SLC7A11-mut + miR-515-5p mimics was indifferent from that of cells transfected with SLC7A11-mut + miR-515-5p mimics (Figure 4B). qRT-PCR showed that, in KGN and SVOG cells, the expression level of SLC7A11 in miR-NC mimic transfected cells was significantly higher than that in miR-515-5p mimic transfected cells (Figure 4C). To further verify the binding of SLC7A11 to miR-515-5p, we performed Ago2-RIP detection in KGN cells. The results showed that circRHBG, SLC7A11, and miR-515-5p were mainly enriched in the Ago2 complex (Figure 4D). In addition, knocking down the expression of circRHBG significantly reduced the enrichment of circRHBG in the Ago2 complex, while increased the enrichment of SLC7A11 in the Ago2 complex (Figure 4D), indicating that circRHBG competes with SLC7A11 to bind to miR-515-5p.
Knockdown of circRHBG promotes ferroptosis in PCOS
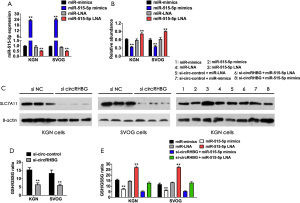
Many studies have confirmed that the overexpression of SLC7A11 promotes tumor growth by inhibiting iron death (24). qRT-PCR showed that, in KGN and SVOG cells, miR-515-5p mimics significantly increased the expression of miR-515-5p, and PBS-formulated LNA-modified oligonucleotide (LNA-anti-miR) effectively antagonized miR-515-5p in these 2 cells (Figure 5A). CCK-8 assay results showed that, in KGN and SVOG cells, cells with a high expression of miR-515-5p (transfected with miR-515-5p mimics) showed higher cell viability than those with a low expression of miR- 515-5p cells (transfected with LNA-anti-miRs) (Figure 5B). Western blot results also confirmed that interference with circRHBG expression in KGN and SVOG can significantly upregulate the expression of SLC7A11, and simultaneous transfection of si-circRHBG and miR-515-5p LNA can reverse the changes in SLC7A11 expression (Figure 5C). In addition, we found that circRHBG knockdown leads to a decrease in the GSH/GSSG ratio (Figure 5D), and LNA-mediated miR-515-5p silencing can reverse the change in the GSH/GSSG ratio (Figure 5E). The decrease in the GSH/GSSG ratio can lead to the inactivation of GPX4, a key molecule that regulates the ferroptosis pathway, and reduce lipid peroxides (25). Therefore, it can be concluded that circRHBG knockdown promotes ferroptosis in PCOS cells through the circRHBG/miR-515-5p/SLC7A11 axis.
Discussion
Many studies have found that a variety of miRNA and long-chain non-coding RNA (lncRNA) can regulate the progress of PCOS. However, few studies have investigated the role of circRNAs in PCOS (23). So far, four studies have investigated the difference in circRNA between PCOS patients and controls, in which hsa_circ_0083952, hsa_circ_0082709, hsa_circ_0002425, and hsa_circ_0015168, etc. have been confirmed to be dysregulated in PCOS patients (22,26-28). In the present study, we determined the expression profile of circRNA in PCOS and found that the expression of circRHBG was upregulated in PCOS granulosa cells. A series of cell experiments showed that knockdown of circRHBG expression enhanced the proliferation ability of KGN and SVOG cells. These results indicate that circRHBG plays a vital role in the progression of PCOS and could be a biomarker and therapeutic target for PCOS diagnosis.
The mechanism of ceRNA has become a research hotspot in recent years. It refers to the phenomenon that lncRNA, circRNA, and even pseudogenes regulate the expression of miRNA target genes through competition for miRNA binding sites (24). circRNA has been shown to regulate gene expression in a variety of diseases through the ceRNA mechanism (11,25). In the present study, we tested circRNA expression profiles in PCOS granulosa cells and then conducted a bioinformatics analysis. The findings indicated that the research molecule, circRHBG, contained miR-515-5p miRNA response element. We then performed functional tests to confirm the direct relationship between circ0097009 and miR-515-5p. Previous studies have found that miR-515-5p plays a regulatory role in a variety of diseases. Zhang et al. confirmed that miR-515-5p exerts a tumor-suppressor effect by targeting TRIP13 in prostate cancer (29). In addition, miR-515p has been shown to be involved in the epithelial-mesenchymal transition of epithelial cells in the process of smoking-induced airway remodeling (30). Pardo et al. also discussed the role of miR-515-5p/MARK4 in cancer cell migration (31). However, the role of miR-515-5p in the progression of PCOS has not been well studied. In the present study, we found that both circRHBG and SLC7A11 can directly bind to miR-515-5p, which indicates that circRHBG may act as a miR-515-5p sponge to regulate the expression of SLC7A11 through the ceRNA mechanism. Our research has progressively confirmed through multiple experiments that circRHBG acts as the ceRNA of SLC7A11 in PCOS and as a sponge for miR-1261. As mentioned earlier, the bioinformatics analysis showed that both 3'-UTR and circRHBG of SLC7A11 contained miR-515-5p binding sites. Second, the dual-luciferase reporter experiment and MS2bp-MS2bs-based RIP detection confirmed this prediction. Third, knockdown of the circRHBG reduced the expression of SLC7A11. Finally, miR-515-5pLNA reversed the effect of circRHBG knockdown on SLC7A11. These findings indicate that circRHBG acts as a ceRNA to regulate the expression of SLC7A11 through sponge miR-515-5p.
Ferroptosis is a novel type of cell death discovered in recent years. The process of cell death is usually characterized by a large amount of iron accumulation and lipid peroxidation (32). Iron metabolism is involved in endocrine diseases, including PCOS (33,34), while its underlying mechanism remains unclear. SLC7A11, the cystine/glutamate antiporter, functions to transport cysteine for glutathione synthesis and antioxidation, which is an upstream regulator of the ferroptosis pathway and can inhibit ferroptosis when its expression is upregulated (35). Ferroptosis is a recently discovered form of regulated cell death (36). It is characterized by the accumulation of lipid peroxidation products and reactive oxygen species (ROS) produced by iron metabolism, which can be pharmacologically inhibited by iron chelating agents and lipid peroxidation inhibitors (37). Our results showed that circRHBG knockdown reduced the expression level of SLC7A11, indicating that circRHBG is involved in the progression of PCOS and may play a role in regulating SLC7A11 to affect the ferroptosis pathway. In addition, we also found that circRHBG knockdown resulted in a decrease in the GSH/GSSG ratio, which leads to the inactivation of GPX4, a key molecule that regulates the ferroptosis pathway, and reduces lipid peroxides GPX4 (38). Studies have shown that silencing GPX4 makes tumor cells sensitive to ROS-induced cell death (39). Therefore, based on these findings, we conclude that the high expression of circRHBG inhibits iron death in PCOS through the Xc- and GPX4 pathways.
The findings of the present study confirmed the carcinogenic effect of circRHBG in PCOS, which plays a regulatory role in the circRHBG/miR515-5p/SLC7A11 axis. The establishment of the ceRNA network, circRHBG/miR515-5p/SLC7A11, in the PCOS model may provide new diagnostic molecular markers and therapeutic targets for PCOS.
Acknowledgments
Funding: This work was supported by National Natural Science Foundation of China (No. 81901445) and Guangdong Provincial Key Laboratory of Reproductive Medicine (No. 2020B1212060029).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-4174
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-4174
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-4174). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All patients provided signed written informed consent, and all experiments were performed under the approval of the local ethics committee of the First Affiliated Hospital of Sun Yat-Sen University ([2021]399-1). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Escobar-Morreale HF. Polycystic ovary syndrome: definition, aetiology, diagnosis and treatment. Nat Rev Endocrinol 2018;14:270-84. [Crossref] [PubMed]
- . Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril 2004;81:19-25. [Crossref]
- Azziz R, Carmina E, Dewailly D, et al. Positions statement: criteria for defining polycystic ovary syndrome as a predominantly hyperandrogenic syndrome: an Androgen Excess Society guideline. J Clin Endocrinol Metab 2006;91:4237-45. [Crossref] [PubMed]
- Asunción M, Calvo RM, San Millán JL, et al. A prospective study of the prevalence of the polycystic ovary syndrome in unselected Caucasian women from Spain. J Clin Endocrinol Metab 2000;85:2434-8. [Crossref] [PubMed]
- Azziz R, Woods KS, Reyna R, et al. The prevalence and features of the polycystic ovary syndrome in an unselected population. J Clin Endocrinol Metab 2004;89:2745-9. [Crossref] [PubMed]
- Cooper HE, Spellacy WN, Prem KA, et al. Hereditary factors in the Stein-Leventhal syndrome. Am J Obstet Gynecol 1968;100:371-87. [Crossref] [PubMed]
- Givens JR. Familial polycystic ovarian disease. Endocrinol Metab Clin North Am 1988;17:771-83. [Crossref] [PubMed]
- Escobar-Morreale HF, Samino S, Insenser M, et al. Metabolic heterogeneity in polycystic ovary syndrome is determined by obesity: plasma metabolomic approach using GC-MS. Clin Chem 2012;58:999-1009. [Crossref] [PubMed]
- Wu G, Xia J, Yang Z, et al. CircASPH promotes KGN cells proliferation through miR-375/MAP2K6 axis in Polycystic Ovary Syndrome. J Cell Mol Med 2020; Epub ahead of print. [Crossref] [PubMed]
- Xia H, Zhao Y. miR-155 is high-expressed in polycystic ovarian syndrome and promotes cell proliferation and migration through targeting PDCD4 in KGN cells. Artif Cells Nanomed Biotechnol 2020;48:197-205. [Crossref] [PubMed]
- Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA 2014;20:1829-42. [Crossref] [PubMed]
- Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell 2013;51:792-806. [Crossref] [PubMed]
- Liu Y, Su H, Zhang J, et al. Back-spliced RNA from retrotransposon binds to centromere and regulates centromeric chromatin loops in maize. PLoS Biol 2020;18:e3000582 [Crossref] [PubMed]
- Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010;465:1033-8. [Crossref] [PubMed]
- Du WW, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 2016;44:2846-58. [Crossref] [PubMed]
- Burd CE, Jeck WR, Liu Y, et al. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 2010;6:e1001233 [Crossref] [PubMed]
- Shan K, Liu C, Liu BH, et al. Circular Noncoding RNA HIPK3 Mediates Retinal Vascular Dysfunction in Diabetes Mellitus. Circulation 2017;136:1629-42. [Crossref] [PubMed]
- Guarnerio J, Bezzi M, Jeong JC, et al. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell 2016;165:289-302. [Crossref] [PubMed]
- Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell 2017;66:22-37.e9. [Crossref] [PubMed]
- Sang Q, Yao Z, Wang H, et al. Identification of microRNAs in human follicular fluid: characterization of microRNAs that govern steroidogenesis in vitro and are associated with polycystic ovary syndrome in vivo. J Clin Endocrinol Metab 2013;98:3068-79. [Crossref] [PubMed]
- Roth LW, McCallie B, Alvero R, et al. Altered microRNA and gene expression in the follicular fluid of women with polycystic ovary syndrome. J Assist Reprod Genet 2014;31:355-62. [Crossref] [PubMed]
- Wang LP, Peng XY, Lv XQ, et al. High throughput circRNAs sequencing profile of follicle fluid exosomes of polycystic ovary syndrome patients. J Cell Physiol 2019; Epub ahead of print. [Crossref] [PubMed]
- Mu L, Sun X, Tu M, et al. Non-coding RNAs in polycystic ovary syndrome: a systematic review and meta-analysis. Reprod Biol Endocrinol 2021;19:10. [Crossref] [PubMed]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014;505:344-52. [Crossref] [PubMed]
- Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell 2018;71:428-42. [Crossref] [PubMed]
- Che Q, Liu M, Xu J, et al. Characterization of circular RNA expression profiles in cumulus cells from patients with polycystic ovary syndrome. Fertil Steril 2019;111:1243-1251.e1. [Crossref] [PubMed]
- Ma Z, Zhao H, Zhang Y, et al. Novel circular RNA expression in the cumulus cells of patients with polycystic ovary syndrome. Arch Gynecol Obstet 2019;299:1715-25. [Crossref] [PubMed]
- Zhang C, Liu J, Lai M, et al. Circular RNA expression profiling of granulosa cells in women of reproductive age with polycystic ovary syndrome. Arch Gynecol Obstet 2019;300:431-40. [Crossref] [PubMed]
- Zhang X, Zhou J, Xue D, et al. MiR-515-5p acts as a tumor suppressor via targeting TRIP13 in prostate cancer. Int J Biol Macromol 2019;129:227-32. [Crossref] [PubMed]
- Ma H, Lu L, Xia H, et al. Circ0061052 regulation of FoxC1/Snail pathway via miR-515-5p is involved in the epithelial-mesenchymal transition of epithelial cells during cigarette smoke-induced airway remodeling. Sci Total Environ 2020;746:141181 [Crossref] [PubMed]
- Pardo OE, Castellano L, Munro CE, et al. miR-515-5p controls cancer cell migration through MARK4 regulation. EMBO Rep 2016;17:570-84. [Crossref] [PubMed]
- Li J, Cao F, Yin HL, et al. Ferroptosis: past, present and future. Cell Death Dis 2020;11:88. [Crossref] [PubMed]
- Yin J, Hong X, Ma J, et al. Serum Trace Elements in Patients With Polycystic Ovary Syndrome: A Systematic Review and Meta-Analysis. Front Endocrinol (Lausanne) 2020;11:572384 [Crossref] [PubMed]
- Escobar-Morreale HF. Iron metabolism and the polycystic ovary syndrome. Trends Endocrinol Metab 2012;23:509-15. [Crossref] [PubMed]
- Koppula P, Zhuang L, Gan B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell 2021;12:599-620. [Crossref] [PubMed]
- Xie Y, Hou W, Song X, et al. Ferroptosis: process and function. Cell Death Differ 2016;23:369-79. [Crossref] [PubMed]
- Stockwell BR, Friedmann Angeli JP, Bayir H, et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017;171:273-85. [Crossref] [PubMed]
- Seibt TM, Proneth B, Conrad M. Role of GPX4 in ferroptosis and its pharmacological implication. Free Radic Biol Med 2019;133:144-52. [Crossref] [PubMed]
- Kinowaki Y, Kurata M, Ishibashi S, et al. Glutathione peroxidase 4 overexpression inhibits ROS-induced cell death in diffuse large B-cell lymphoma. Lab Invest 2018;98:609-19. [Crossref] [PubMed]
(English Language Editor: R. Scott)


