
Co-mutations of epidermal growth factor receptor and BRAF in Chinese non-small cell lung cancer patients
Introduction
The incidence and mortality of lung cancer rank first among all types of cancers worldwide (1). Non-small cell lung cancer (NSCLC) is the major histological subtype of lung cancer, accounting for approximately 75–80% of all cases (2). Epidermal growth factor receptor (EGFR) is the most frequent somatic mutation driver gene in NSCLC, detected in ~30–40% of Asian patients (3). With the development of precision medicine, tyrosine kinase inhibitors (TKIs) have become the standard drug treatment for NSCLC patients harboring EGFR somatic mutations and have greatly improved overall survival. However, acquired resistance (AR) to EGFR-TKIs always occurs after targeted therapy. Different mechanisms have been discovered to be responsible for AR, including on-target (EGFR-dependent) and off-target (EGFR-independent) (4). Such as EGFR T790M mutation (~50–60%) after first- or second-generation EGFR TKIs, MET amplification (~20%), and transformation to small-cell lung cancer (SCLC) (~5–10%), among others (5). BRAF is another driver gene found in NSCLC. The frequency of BRAF mutations is relatively low (~2–5%) (6-8). BRAF alterations were found in 4.4% of Chinese NSCLC patients (N=1,200) (9). BRAF mutations include V600E, promoting several fold kinase hyperactivation; non-V600E activating mutations, rearrangements, N-terminal deletions (NTDs), kinase domain duplications (KDDs), and fusions, resulting in constitutive activation of BRAF and downstream ERK signalling (10-12). Dabrafenib and trametinib are approved for the management of advanced NSCLCs that harbor BRAF V600E mutations.
EGFR and BRAF mutations are normally mutually exclusive, as the coexistence of EGFR and BRAF somatic mutations are uncommon in NSCLC patients. The frequency of EGFR and BRAF co-mutation in the western population is around 0.97% [cBioPortal database (http://www.cbioportal.org)]. With the accumulation of NSCLC patients, some studies have reported the existence of actionable BRAF mutations in EGFR-mutated NSCLC patients. A study on 5,125 Chinese NSCLC patients found that only 2 of them harbored both EGFR and BRAF mutations (13). Another study on patients with AR to EGFR-TKIs detected BRAF mutations in 2 patients (G469A and V600E), and cell line experiments demonstrated that BRAF V600E could cause resistance to erlotinib (14). These studies highlight the possibility that BRAF mutations are likely to be another emerging mechanism of AR to EGFR-TKIs. In a study of 326 non-squamous NSCLC patients, 240 (73.6%) had EGFR mutations, and of these 240 patients with EGFR determination, 2.9% had BRAF mutations (15). BRAF was shown to be altered in 4.5% of western NSCLC patients, and 37.4% (n=397) had BRAF V600E, 38% had BRAF non-V600E activating mutations, and 18% had BRAF inactivating mutations. Rearrangements were observed at a frequency of 4.3% (10).
Limited to the number of patients carrying both EGFR and BRAF mutations, the influence of different BRAF mutations on EGFR-TKIs is not yet clear. This study aims to determine the incidence of various EGFR and BRAF co-mutations in Chinese NSCLC patients and the influence of different types of BRAF mutations (including short variants, copy number changes, and rearrangements) on EGFR-TKI-treated Chinese NSCLC patients. We present the following article in accordance with the STROBE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-3570).
Methods
Sample collection
We retrospective analysed from 13,976 Chinese NSCLC patient samples that formalin-fixed, paraffin-embedded (FFPE) tumor samples and matched blood samples were collected and prepared according to standard procedures. All cases were diagnosed with lung cancer according to the World Health Organization criteria based on hematoxylin and eosin staining reviewed by experienced pathologists, including lung adenocarcinoma, squamous cell lung carcinoma, and adenosquamous carcinoma of the lung, among other types. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics board of Guangdong Second Provincial General Hospital (No.: GZR-2020-KT-39-01) and informed consent was taken from all the patients.
Next-generation sequencing (NGS)
Genomic alterations of patients were detected by the tissue-based 450 genes panel assay for FFPE samples with paired blood as normal control, and circulating tumor DNA (ctDNA) based for 329 or 18 genes panel. All samples were sequenced in a College of American Pathologists (CAP) accredited and Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory (OrigiMed, Shanghai, China).
At least 50 ng of cancer tissue DNA was extracted from each 40-mm FFPE tumor sample using a DNA Extraction Kit (QIAamp DNA FFPE Tissue Kit) according to the manufacturer’s protocol. All coding exons of 450 key cancer-related genes and selected introns of 36 genes commonly rearranged in solid tumors were incorporated into the custom hybridization capture panel (YuansuTM, OrigiMed) (16). Libraries were constructed and each diluted to 1.05 nM and then sequenced with a mean coverage of 900× for tissue samples (minimum 700×) and 300× for matched blood samples on an Illumina NextSeq-500 Platform (Illumina Incorporated, San Diego, CA, USA).
Bioinformatics analysis, variant identification, and annotation
Genomic alterations including single nucleotide variants (SNVs), short and long insertions/deletions (Indels), copy number variations (CNVs), gene rearrangements, and fusions were subjected to advanced analysis. First, reads were aligned to a human genome reference sequence (hg19) by Burrows-Wheeler-Alignment (BWA) (17), and polymerase chain reaction (PCR) duplicates were removed using Picard (available online: https://broadinstitute.github.io/picard/). Second, SNVs and short Indels were identified by MuTect (18) after quality recalibration and realignment using GATK (Broad Institute, Cambridge, MA, USA) and an in-house pipeline. Short Indels were then calibrated using the results from Pindel (19). The log-ratio per region of each gene was calculated, and customized algorithms were used to detect copy number changes. Tumor cellularity was estimated by allele frequencies of sequenced SNPs. A customized algorithm was developed to detect gene rearrangements and long Indels. Reliable somatic alterations were detected in the raw data by comparing tumor tissues with matched blood control samples. At minimum, 5 reads and a minimum variant allele frequency of 1% were required to support alternative calling. For the calling of gene rearrangements and fusions, aligned reads with an abnormal insert size of over 2,000 or 0 bp were collected and used as discordant reads. Next, the discordant reads with a distance less than 500 bp formed clusters that were further assembled to identify potential rearrangement breakpoints. The breakpoints were confirmed by the BLAST-like alignment tool and the resulting chimeric gene candidates were annotated.
Statistical analysis
The variants were divided into 4 tiers after identification (20): Tier I, variants with strong clinical significance; Tier II, variants with potential clinical significance; Tier III, variants of unknown clinical significance; and Tier IV, variants deemed benign or likely benign. Tier I and Tier II were considered clinically significant mutations, which is the focus of future analysis. IBM SPSS Statistics (Version 20.0; IBM Corp, Armonk, NY) was used for statistical analysis. For all test, P<0.05 was defined as statistically significant.
Results
A total of 127 patients harboring both EGFR and BRAF mutations were included in this study. Of these patients, 93 harbored clinically significant mutations of both EGFR and BRAF. The remaining 34 patients had mutations of uncertain significance of either EGFR or BRAF. We aimed to explore the association between BRAF mutations and EGFR-TKI AR, and focused on the analysis of 93 patients harboring both EGFR and BRAF clinically significant mutations.
Demographic and clinicopathological data of the patients
The demographics and clinicopathological data of patients in the cohort are summarized in Table 1. The median age of patients at the time of sampling was approximately 60 years (range, 33–82 years), and females were moderately overrepresented compared with males (56% of patients were female). There were approximately equal numbers of male and female patients older than 60. However, female patients were overrepresented in the ≤60 age group (66% vs. 34%), which is slightly different to Chinese lung cancer patients (9). Regarding histological subtypes, most patients had lung adenocarcinoma (95%), while all other patients had squamous cell lung carcinoma. Patients were classified into main clinical stages (I–IV) according to both pathology and medical history following the American Journal of Critical Care Cancer Staging Manual (version 8; Table 1). More than half of the patients were late stage (III–IV).
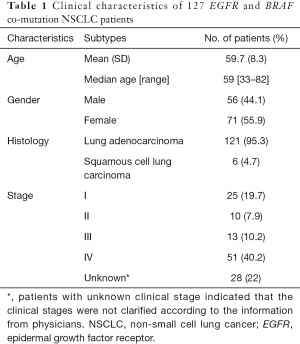
Full table
Profiling of 18 actionable genes of EGFR and BRAF co-mutation NSCLC patients
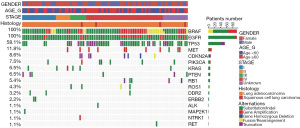
We analyzed 18 actionable genes of the 93 NSCLC patients, including AKT1, ALK, BRAF, CDKN2A, DDR2, EGFR, ERBB2, KRAS, MAP2K1, MET, NRAS, NTRK1, PIK3CA, PTEN, RB1, RET, ROS1, and TP53. Profiling of the 18 actionable genes of the 93 NSCLC patients was conducted as shown in Figure 1. TP53 was the most frequently mutated gene in BRAF and EGFR co-mutated patients, accounting for approximately 58% (N=54/93). MET active mutations (amplification and exon 14 skipping) accounted for 12% (N=11/93). CDKN2A mutations accounted for 8.6% (N=8/93) and PIK3CA mutations accounted for 7.5% (N=7/93). CDKN2A and PIK3CA mutations were more frequently observed in late-stage (III–IV) EGFR and BRAF co-mutated patients.
Distribution of EGFR mutations in NSCLC patients
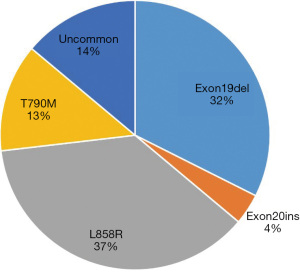
In regards to EGFR mutations, which excluded amplification of EGFR, 86% of EGFR-mutant patients harbored hotspots, including L858R (37%), exon 19 deletion (32%), T790M (13%), and exon 20 insertions (4%). Figure 2 shows the profile of EGFR genomic alterations. Moreover, uncommon mutations accounted for 14% of EGFR mutations. Uncommon EGFR mutations were defined as mutations other than L858R, exon 19del, and exon 20ins, including KDD (exon18_exon25dup) and G719/S768 mutation.
Distribution of EGFR and BRAF subtypes of EGFR and BRAF co-mutation NSCLC patients
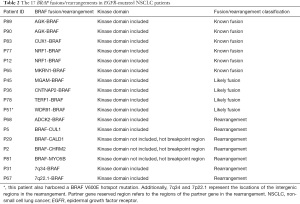
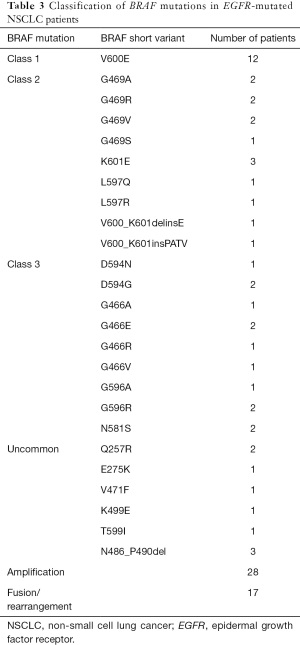
BRAF mutations included short variants and fusions. Short variants of BRAF were divided into 3 classes: RAS-independent and signal as active monomers (class 1), constitutively active dimers (class 2), and impaired kinase activity or are kinase-dead (class 3) (21). BRAF fusions/rearrangements were divided into 3 types depending on different partner genes and breakpoints. including: (I) likely fusion, refer to the 5' region of a novel partner gene with the kinase domain-containing 3' region of BRAF; (II) known fusion, refer to the 5' region of a known partner gene with the kinase domain-containing 3' region of BRAF; (III) Rearrangements, any other forms were classified as rearrangements. Approximately 18% of patients (N=17/93) with clinically significant EGFR mutations were found to have combined BRAF fusions/rearrangements (Table 2).
Full table
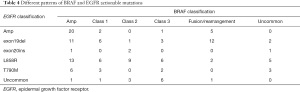
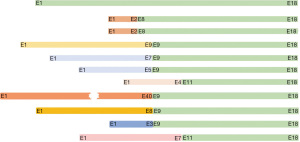
The number of short variants of BRAF was 12 for class 1, 14 for class 2, and 13 for class 3. There were 9 uncommon active mutations. Also, there were 28 patients harboring BRAF amplification, and 17 patients had BRAF fusion and rearrangement (Table 3). Of these 17 patients, EGFR mutations were mainly exon19del (N=12/17) (Table 4). BRAF fusion was more likely detected in EGFR exon19del patients compared with non-exon19del patients (P value =0.015). There have been 6 known fusions reported in the literature, including AGK-BRAF in sporadic pediatric papillary thyroid carcinoma (22), CUX1-BRAF in metanephric adenoma (23), NRF1-BRAF in urothelial carcinoma (24), and MKRN1-BRAF in thyroid carcinomas (25). Additionally, there were 4 BRAF likely fusions and 7 BRAF rearrangements, and 9 of them were with a novel partner. NTDs and KDDs were not detected in our cohort. The breakpoints of BRAF fusion/rearrangement were located at the known hot regions (intron7/8/10) (Figure 3).
Full table
Full table
EGFR mutations found in patients with BRAF rearrangements were mainly exon19del (N=12/17). Two patients carried EGFR L858R and another 2 had EGFR amplification. EGFR KDD was also found in 1 patient. EGFR T790M was not found in any patient with BRAF fusion/rearrangement. Furthermore, 28 of 93 patients (about 30%) with significant EGFR mutations harbored BRAF amplification. Of these patients, EGFR amplification, L858R, exon19del, and T790M accounted for around 71% (N=20/28), 43% (N=12/28), 46% (N=13/28), and 21% (N=6/28), respectively. EGFR amplification was the most frequent mutation type with BRAF amplification.
There were 2 patients with emerging BRAF fusions following progression on TKI therapy (Figure 4): (I) EGFR exon 19del was identified in patient A. The patient was started on gefitinib in January 2017 and disease progression occurred 1 year later. A repeat lung biopsy detected EGFR T790M, and osimertinib treatment was used for about year and a half. Unfortunately, the disease progressed again after osimertinib treatment. A lung biopsy revealed that this patient harbored EGFR exon19del, EGFR amplification, and CUX1-BRAF fusion, and T790M disappeared. (II) Patient B was similar to patient A. The patient experienced rapid disease progression after osimertinib treatment and harbored NRF1-BRAF fusion.
Discussion
In this study, we present, to our knowledge, the largest cohort of BRAF and EGFR co-mutation Chinese NSCLC patients. A total of 127 Chinese NSCLC patients harboring co-mutations of EGFR and BRAF mutations were enrolled in this study. BRAF fusion was more likely detected in EGFR exon19del patients compared with non-exon19del patients (P value =0.015). Aside from BRAF V600E, class II/III and BRAF fusions were found, which provides clues for investigating the resistance mechanisms of EGFR-TKIs in the future.
Pathological activation of the RAS/RAF/MEK/ERK (MAPK) pathway is observed across multiple tumor types, and BRAF alterations in lung cancer can be targeted by MEK inhibitors or pan-RAF inhibitors. BRAF mutations were found in 4–5% of NSCLC patients. The US Food and Drug Administration (FDA) has approved combined dabrafenib and trametinib therapy for metastatic NSCLC with BRAF V600E mutation. EGFR mutations, the most common alterations in lung cancer, account for the majority of druggable targets in lung adenocarcinoma. Over the past decades, the optimization of EGFR inhibitors has revolutionized the treatment options for patients suffering from this disease (26). For lung cancer patients with EGFR exon 19 deletions or an exon 21 Leu858Arg mutation, the standard first-line treatments are first-generation (gefitinib, erlotinib) or second-generation (afatinib) TKIs. EGFR-TKIs improve response rates, time to progression, and overall survival. Unfortunately, patients with EGFR-mutant lung cancer develop disease progression after a median of 10 to 14 months on EGFR-TKIs. Different mechanisms of AR to first-generation and second-generation EGFR-TKIs have been reported. Optimal treatments for the various mechanisms of AR have not yet been clearly defined, except for the T790M mutation. Osimertinib has been approved for patients with T790M-positive NSCLC with AR to EGFR-TKIs. For other TKI resistance mechanisms, combination therapy may be considered (27).
Usually, combined BRAF and EGFR mutations are rare in NSCLC. The frequency of EGFR and BRAF co-mutation was 0.91% in Chinese NSCLC patients, similar to Western patients. Among the 127 patients with both EGFR and BRAF mutations, 93 of them harbored clinically significant mutations. The remaining 34 patients had mutations of uncertain significance of either EGFR or BRAF. Approximately 18% of patients (N=17/93) with significant EGFR mutations were detected to have fusions/rearrangements of the BRAF gene. Of these 17 patients, EGFR mutations were mainly exon19del. Two patients carried EGFR L858R and another 2 had EGFR amplification. EGFR KDD (exon18_exon25dup) was also found in 1 patient. In addition, EGFR T790M, as the most TKI-resistant mutation, was not found in any patient with BRAF fusion/rearrangement. This may indicate that BRAF fusion is an AR mechanism against osimertinib, similar to EGFR C797S after osimertinib treatment in T790M patients (28).
The prognosis of EGFR-TKI treatment can vary according to different BRAF actionable mutations. Multiple genetic mechanisms have been identified in EGFR-mutant lung cancers as mediators of AR to EGFR-TKIs. The most common mechanisms of AR include secondary EGFR mutation, MET amplification, and histologic transformation (29,30). We also found that BRAF fusion was more likely detected in patients with EGFR exon19del. Additionally, several novel BRAF fusion partners were detected. Aside from BRAF V600E, class II/III were found, which provides clues for investigating the resistance mechanisms of EGFR-TKIs in the future. Although BRAF fusion may seem to be an obvious therapeutic target, the US FDA-approved BRAF inhibitors have not been effective against BRAF fusions. Regarding off-target (EGFR independent) resistance mechanisms, combinations of EGFR TKIs with different drugs (including other TKIs, monoclonal antibodies, chemotherapy and vaccines) are currently under investigation (4). Several clinical trials was ongoing including Biomarker-driven approaches, such as combination of osimertinib and the MET TKI savolitinib for MET amplification and the objective response rate (ORR) was 30% in patients previously treated with third-generation EGFR TKIs, with a median PFS of 5.4 months (31). Also the combination therapy has been tried for fusion caused resistance of EGFR TKI, there was 2 NSCLC patients with EGFR-mutant and RET-fusion was treated with osimertinib and BLU-667 and was well tolerated with rapid radiographic response (32). Among melanomas that harbor BRAF fusions, response to trametinib has been described, which indicates that NSCLC tumors that harbor BRAF fusions may also benefit from monotherapy with MEK inhibitors (33,34). These findings revealed that combined inhibition of EGFR and MEK (with osimertinib and trametinib) or BRAF (with a pan-RAF inhibitor) are potential therapeutic strategies that should be explored. Due to the limitations of the number of samples, more follow up data about therapy response information is needed for further research.
Acknowledgments
The authors would like to thank the patients for providing written informed consent for publication and all the research staff involved in this study. The authors thank OrigiMed for next-generation sequencing technical support and scientific comments.
Funding: This work was supported partially by Wujieping Foundation (320.6750.19092-25). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-3570
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-3570
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-3570). Dr. JH and Dr. KW are from OrigiMed Co., Ltd. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics board of Guangdong Second Provincial General Hospital (No.: GZR-2020-KT-39-01) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Zheng H, Zhan Y, Liu S, et al. The roles of tumor-derived exosomes in non-small cell lung cancer and their clinical implications. J Exp Clin Cancer Res 2018;37:226. [Crossref] [PubMed]
- Passaro A, Pochesci A, Spitaleri G, et al. Afatinib for the first-line treatment of patients with metastatic EGFR-positive NSCLC: a look at the data. Expert Rev Clin Pharmacol 2016;9:1283-8. [Crossref] [PubMed]
- Passaro A, Jänne PA, Mok T, et al. Overcoming therapy resistance in EGFR-mutant lung cancer. Nat Cancer 2021;2:377-91. [Crossref]
- Remon J, Steuer CE, Ramalingam SS, et al. Osimertinib and other third-generation EGFR TKI in EGFR-mutant NSCLC patients. Ann Oncol 2018;29:i20-7. [Crossref] [PubMed]
- Marchetti A, Felicioni L, Malatesta S, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol 2011;29:3574-9. [Crossref] [PubMed]
- Pratilas CA, Hanrahan AJ, Halilovic E, et al. Genetic predictors of MEK dependence in non-small cell lung cancer. Cancer Res 2008;68:9375-83. [Crossref] [PubMed]
- Paik PK, Arcila ME, Fara M, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol 2011;29:2046-51. [Crossref] [PubMed]
- Wen S, Dai L, Wang L, et al. Genomic Signature of Driver Genes Identified by Target Next-Generation Sequencing in Chinese Non-Small Cell Lung Cancer. Oncologist 2019;24:e1070-81. [Crossref] [PubMed]
- Sheikine Y, Pavlick D, Klempner SJ, et al. BRAF in Lung Cancers: Analysis of Patient Cases Reveals Recurrent BRAF Mutations, Fusions, Kinase Duplications, and Concurrent Alterations. JCO Precis Oncol 2018; [Crossref] [PubMed]
- Dankner M, Rose AAN, Rajkumar S, et al. Classifying BRAF alterations in cancer: new rational therapeutic strategies for actionable mutations. Oncogene 2018;37:3183-99. [Crossref] [PubMed]
- Karoulia Z, Gavathiotis E, Poulikakos PI. New perspectives for targeting RAF kinase in human cancer. Nat Rev Cancer 2017;17:676-91. [Crossref] [PubMed]
- Li S, Li L, Zhu Y, et al. Coexistence of EGFR with KRAS, or BRAF, or PIK3CA somatic mutations in lung cancer: a comprehensive mutation profiling from 5125 Chinese cohorts. Br J Cancer 2014;110:2812-20. [Crossref] [PubMed]
- Ohashi K, Sequist LV, Arcila ME, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci U S A 2012;109:E2127-33. [Crossref] [PubMed]
- Martín Martorell P, Huerta M, Compañ Quilis A, et al. Coexistence of EGFR, KRAS, BRAF, and PIK3CA Mutations and ALK Rearrangement in a Comprehensive Cohort of 326 Consecutive Spanish Nonsquamous NSCLC Patients. Clin Lung Cancer 2017;18:e395-402. [Crossref] [PubMed]
- Cao J, Chen L, Li H, et al. An Accurate and Comprehensive Clinical Sequencing Assay for Cancer Targeted and Immunotherapies. Oncologist 2019;24:e1294-302. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009;25:1754-60. [Crossref] [PubMed]
- Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213-9. [Crossref] [PubMed]
- Ye K, Schulz MH, Long Q, et al. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 2009;25:2865-71. [Crossref] [PubMed]
- Li MM, Datto M, Duncavage EJ, et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn 2017;19:4-23. [Crossref] [PubMed]
- Yao Z, Yaeger R, Rodrik-Outmezguine VS, et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature 2017;548:234-8. [Crossref] [PubMed]
- Cordioli MI, Moraes L, Carvalheira G, et al. AGK-BRAF gene fusion is a recurrent event in sporadic pediatric thyroid carcinoma. Cancer Med 2016;5:1535-41. [Crossref] [PubMed]
- Ding Y, Wang C, Li X, et al. Novel clinicopathological and molecular characterization of metanephric adenoma: a study of 28 cases. Diagn Pathol 2018;13:54. [Crossref] [PubMed]
- Isaacson AL, Guseva NV, Bossler AD, et al. Urothelial carcinoma with an NRF1-BRAF rearrangement and response to targeted therapy. Cold Spring Harb Mol Case Stud 2019;5:a003848 [Crossref] [PubMed]
- Chu YH, Wirth LJ, Farahani AA, et al. Clinicopathologic features of kinase fusion-related thyroid carcinomas: an integrative analysis with molecular characterization. Mod Pathol 2020;33:2458-72. [Crossref] [PubMed]
- Tumbrink HL, Heimsoeth A, Sos ML. The next tier of EGFR resistance mutations in lung cancer. Oncogene 2021;40:1-11. [Crossref] [PubMed]
- Wu SG, Shih JY. Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Mol Cancer 2018;17:38. [Crossref] [PubMed]
- Jia Y, Yun CH, Park E, et al. Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature 2016;534:129-32. [Crossref] [PubMed]
- Shao Y, Zhong D. Gene Fusions as Acquired Resistance Mechanisms of EGFR-TKI. Zhongguo Fei Ai Za Zhi 2020;23:381-7. [PubMed]
- Vojnic M, Kubota D, Kurzatkowski C, et al. Acquired BRAF Rearrangements Induce Secondary Resistance to EGFR therapy in EGFR-Mutated Lung Cancers. J Thorac Oncol 2019;14:802-15. [Crossref] [PubMed]
- Sequist LV, Han JY, Ahn MJ, et al. Osimertinib plus savolitinib in patients with EGFR mutation-positive, MET-amplified, non-small-cell lung cancer after progression on EGFR tyrosine kinase inhibitors: interim results from a multicentre, open-label, phase 1b study. Lancet Oncol 2020;21:373-86. [Crossref] [PubMed]
- Piotrowska Z, Isozaki H, Lennerz JK, et al. Landscape of Acquired Resistance to Osimertinib in EGFR-Mutant NSCLC and Clinical Validation of Combined EGFR and RET Inhibition with Osimertinib and BLU-667 for Acquired RET Fusion. Cancer Discov 2018;8:1529-39. [Crossref] [PubMed]
- Ross JS, Wang K, Chmielecki J, et al. The distribution of BRAF gene fusions in solid tumors and response to targeted therapy. Int J Cancer 2016;138:881-90. [Crossref] [PubMed]
- Roskoski R Jr. RAF protein-serine/threonine kinases: structure and regulation. Biochem Biophys Res Commun 2010;399:313-7. [Crossref] [PubMed]
(English Language Editor: C. Betlzar)



