Update in genetic susceptibility in melanoma
Introduction
Melanoma is the most aggressive of the common skin cancers, being responsible for 75% of deaths from skin cancer (1). Melanoma incidence is rapidly increasing especially in Caucasian populations (1,2). Although development of melanoma during childhood is rare, it can appear at any age and is the second most diagnosed cancer among patients under 30 years old (3). For this reason, melanoma is one of the cancers with more years of productive life lost (4). If melanoma is diagnosed in its early stages, it can be cured by surgical removal. However, when the diagnosis is delayed, melanoma is the tumor with the highest metastatic capacity, since it increases by 10% per millimeter of thickness. Despite the improvement in survival of metastatic patients thanks to new targeted therapies, diagnosis and treatment of the initial tumor remains the best strategy for dealing with melanoma (5). Thus, the identification of individuals at high risk of developing melanoma is essential to reduce melanoma mortality, as prevention and early detection programs can be implemented.
Melanoma etiology is complex and heterogeneous as it involves environmental, phenotypic and genetic risk factors. The main environmental risk factor for melanoma is the exposure to ultraviolet radiation (UVR) (6). UVR causes DNA damage through the formation of pyrimidine dimers, photoproducts, gene mutations, oxidative stress, inflammation and immunosuppression, favoring the carcinogenic process (7). UVR has been widely demonstrated to be implicated in nevo-genesis and melanoma-genesis. UVR can induce clinical changes (increased pigmentation, scale formation and erythema) as well as dermoscopic changes in pigmentation (changes in size and number of globules and dots, regression features such as bluish gray granules, blurred pigmented network and increased vascularity) (8-10). The use of sunscreen can prevent part of the UVR effects on nevi (11). Furthermore, a 10-year follow-up study showed that the daily use of sunscreen reduces the melanoma detection rate, suggesting that regular sunscreen use may prevent melanoma development (12). However, there are intrinsic risk factors that can predispose to melanoma. Phenotypic characteristics such as red or blond hair, blue or green eyes, fair skin with low tanning ability, freckles, multiple melanocytic nevi (100 or more) or 5 or more atypical nevi are associated with an increased risk to develop melanoma (13,14). Personal history of melanoma increases 5-8% the risk of developing a second melanoma (15,16). Finally, family history of melanoma has been widely associated with an increased melanoma risk (14).
Familial cancer syndromes are recognized for gathering high-risk features, including a cluster of relatives within the family who have the same or similar cancers as the patient, the development of cancer at a young age, the presentation of more than one synchronous or metachronous primary tumor or more than one tumor within a specific group of tumors that are features of a syndrome (17). Approximately 5-10% of melanoma cases occur in a familial context (18). In these families, melanoma susceptibility is inherited following an autosomal dominant inheritance pattern with incomplete penetrance (19). However, multiple primary melanoma patients also have inherited melanoma susceptibility (20).
This review aims to give an overview of the melanoma susceptibility genes known to date and genetic counseling in melanoma.
Melanoma susceptibility genes
High risk genes
Melanoma high risk genes are defined as genes that when mutated in an individual confer a high risk of developing melanoma and are usually associated with multiple melanoma cases within the family. Cyclin-dependent kinase inhibitor 2A (CDKN2A) was the first gene associated with melanoma susceptibility. The CDKN2A gene is located in the 9p21 locus and encodes two tumor suppressor proteins p16INK4A and p14ARF via differential splicing and alternative reading frames. The protein p16INK4A, encoded by the α-transcript (composed by exon 1α, 2 and 3), promotes the arrest of the cell cycle in the G1 phase by inhibiting RB (retinoblastoma protein) phosphorylation through cyclin-dependent kinase 4 (CDK4). The β-transcript (composed by exon 1β, 2 and 3) encodes p14ARF and acts through the p53 pathway inducing the cell cycle arrest or favoring apoptosis (21). Furthermore, both p53 and p16INK4A play an important role on cell damage response and senescence (22). In 1992, Cannon-Albright and colleagues described for the first time 9p21 as a familial melanoma locus thanks to linkage analyses (23). Two years later the first germline mutations in CDKN2A were reported in familial melanoma (24). Many studies have been performed since then trying to investigate the role of CDKN2A in melanoma susceptibility. To date, CDKN2A is the main high risk gene involved in melanoma susceptibility (25). Mutations in that gene are found in around 20% of melanoma-prone families [Figure 1, references (25-39)], but the CDKN2A mutation frequency can range from 5% to 72% depending on the selection criteria used and the geographical areas (26,40). The CDKN2A mutation detection rate increases with the number of cases in the family. Mutations in CDKN2A are also detected in sporadic multiple primary melanoma patients (SMP). The CDKN2A mutation frequency in SMP with at least two primary melanoma is around 9% (20,27,41). As in familial studies, the probability to detect mutations in SMP increases with the number of total primaries. On the other hand, the probability to have CDKN2A mutations in sporadic melanoma patients without personal or familial history of melanoma is about 1% (42). The penetrance for melanoma in CDKN2A carriers varies between geographical areas and increases with age. Bishop and colleagues reported that at the age of 50, the melanoma penetrance for carriers was 13% in Europe, 50% in the US and 32% in Australia, whilst at the age of 80 the penetrance was 58% in Europe, 76% in the US and 91% in Australia (43). Furthermore, individuals carrying CDKN2A germline mutations have an inherited risk to develop other cancer types beyond melanoma. A strong association between CDKN2A germline mutations and pancreatic cancer has been reported (27,40,44-48). Families with CDKN2A mutations also have an increased risk to develop breast, lung and other tobacco-related cancers (27,47,48).
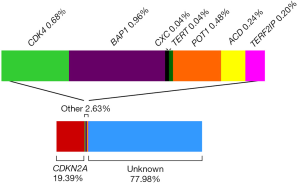
CDK4 was the second high risk melanoma susceptibility gene identified (49). CDK4 is an oncogene located within the 12q14 chromosomal region and encodes a protein that controls cell cycle progression through the G1 phase. To date mutations in this gene have been described in 17 melanoma-prone families and in all of them the mutation affects the same amino acid (Arginine 42) (50). This amino acid is located in the p16INK4A binding domain of the CDK4 protein. Thus, when CDK4 is mutated, p16INK4A cannot inhibit the CDK4 kinase activity resulting in the progression of the cell cycle. CDK4 mutation carriers phenotypically behave similarly to p16INK4A mutation carriers (50). This is consistent with the functional impact that mutations in both genes have at the cellular level, which results in the activation of the same pathway.
Breast cancer 1 (BRCA1) associated protein 1 (BAP1) germline mutations have been associated with a cancer syndrome characterized by the presence of broad tumor types: cutaneous melanoma, uveal melanoma, mesothelioma, renal cell carcinoma (RCC), atypical Spitz tumors, atypical intradermal tumors (MBAITs) and multiple basal cell carcinomas (51-65). However, the whole tumor spectrum associated with germline BAP1 mutations is still unknown. BAP1 is located in the chromosomal region 3p21 and encodes a protein that plays a tumor suppressor role through transcription regulation by chromatin remodeling and the ubiquitin-proteasome system. BAP1 is a deubiquitylase that participates in multi-protein complexes that regulate key pathways including the cell cycle, cell differentiation, cell death, gluconeogenesis and the DNA damage response (54). The frequency of CDKN2A wild-type melanoma-prone families with mutations in BAP1 is not well established, but beyond cutaneous melanoma, families bearing BAP1 mutations seem to be enriched by uveal melanoma, mesothelioma, RCC, other cutaneous tumors and other cancers.
Copy number variants (CNV) assessed genome wide, allowed the identification of a duplicated region on 4q13 segregating with melanoma in one melanoma-prone family (66), suggesting that some melanoma-prone families seem to carry mutations in private genes. The whole duplicated region contains 10 genes, most of them belonging to a family of CXC chemokines, such as melanoma growth-stimulating activity α (CXCL1) and interleukin 8 (IL-8). Both genes have been shown to stimulate melanoma growth in vitro and in vivo (66).
The most recent findings in melanoma susceptibility involve genes that play a role in telomere maintenance. Telomeres consist of tandem nucleotide repeats (TTAGGG) and are located at the ends of chromosomes. The telomerase enzyme, the shelterin protein complex and many other accessory proteins are also comprised in the telomeres. They maintain genomic stability and chromosomal integrity by protecting chromosome ends from degradation, end-to-end fusion, and atypical recombination (67). Telomeres shorten both with age and following exposures associated with cancer risk, such as smoking and ultraviolet (UV) irradiation (68,69). Thus, telomere maintenance processes are natural candidates for explaining carcinogenesis (70). Horn and colleagues identified a germline mutation in the promoter of telomerase reverse transcriptase (TERT) in a melanoma-prone family using multipoint linkage analyses and target-enriched high-throughput sequencing. TERT is located in 5p15 and encodes the catalytic subunit of the telomerase, which is the ribonucleoprotein complex that maintains telomere length (71). Recently, two independent groups identified rare germline variants in protection of telomeres 1 (POT1) in 12 CDKN2A wild-type melanoma-prone families using next generation sequencing (72,73). POT1 is located within the 7q31 chromosomal region and encodes a protein of the telomeric shelterin complex. POT1 plays an important role in telomere maintenance by preventing the inappropriate processing of the exposed chromosome ends, caused by pathways related to DNA damage response, and regulating telomerase function (74). Furthermore, Aoude and colleagues described germline mutations in melanoma-prone families located in two more genes involved in the shelterin complex, ACD and TERF2IP. This study included 510 melanoma-prone families without mutations in the known melanoma susceptibility genes to date. They identified six families with mutations in ACD and four families with TERF2IP mutations. The mutations were segregating with melanoma in the families (75). Overall, the germline mutations in genes that play a role in telomere maintenance (TERT, POT1, ACD and TERF2IP) may explain around 1% of familial melanoma cases, showing the relevance of telomere maintenance in melanoma susceptibility (Figure 1).
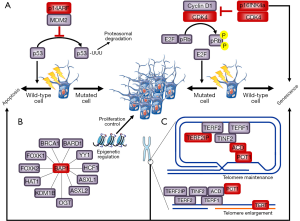
Figure 2 shows the interaction and pathways implicated until now in familial melanoma susceptibility.
Low to moderate risk genes
Melanocortin 1 Receptor (MC1R) is considered a moderate risk gene and its role in melanoma susceptibility has been widely studied. MC1R, located in 16q24, is one of the master regulator genes in human pigmentation and encodes the α melanocyte-stimulating hormone (α-MSH) receptor 1. MC1R is a highly polymorphic gene in the Caucasian population. Variants in MC1R have different functional effects, either at the level of α-MSH binding or cAMP signaling, resulting in changes in the ratio between eumelanin (brown pigment) and pheomelanin (red-yellow pigment, potentially mutagenic) ratio (76). MC1R variants are associated with skin and hair pigmentation and, independently of their phenotypic effect, MC1R variants are associated with an increased risk of developing melanoma (77). When the MC1R function is highly compromised, this usually results in the red hair color phenotype (RHC). The most common MC1R variants have been classified as r variants, when there is a low association with RHC (p.V60L, p.V92M, p.R163Q) and R variants, when they are highly associated with RHC (p.D84E, p.R142H, p.R151C, p.I155T, p.R160W, p.D294H) (78,79). The R variants are those most implicated in melanoma susceptibility. The melanoma risk conferred by R variants varies from two times risk per R allele in the general population to three times risk, in the familial melanoma context. The risk is additive, thus carriers of two R alleles have a 4 to 6 times increased risk than individuals without these variants. Studies assessing the modulator effect of MC1R variants in CDKN2A carriers demonstrate that the presence of MC1R variants increase the melanoma penetrance in CDKN2A carriers (80). The r variant p.R163Q is associated with increased risk of melanoma in high sun exposed geographic areas (81) and with a subtype of melanoma associated with chronic sun damage, lentigo maligna melanoma (82).
Microphthalmia-associated transcription factor (MITF) is also considered a moderate risk gene. Two independent groups identified the rare functional variant in MITF p.E318K (rs149617956) that increases melanoma risk and also predisposes to RCC (83,84). MITF is located in the chromosomal region 3p14 and is a master regulator gene of melanocyte development and differentiation, and it is also associated with melanoma development and progression (85). MITF p.E318K occurs at a conserved SUMOylation position and this variant decreases the number of SUMO-modified MITF forms. As SUMOylation of MITF represses its transcriptional activity, p.E318K increases the MITF transcriptional activity and may result in the up-regulation of distinct sets of genes. Furthermore, this variant promotes invasive and tumorigenic behaviors in melanoma and RCC cells and might favor a phenotypic switch of melanoma cells towards a tumor-initiating cell phenotype (84). The prevalence of p.E318K in melanoma patients ranges from 1.6% to 2.8%, whilst in a control population prevalence of this variant is 0.6% in French and Italian populations and 0.8% in UK and Australian populations (83,84,86). Furthermore, this variant has been associated with phenotypic features such as high nevi count, fair skin and non-blue eye color (83,87,88).
Other genes containing common variants in the population show association with melanoma, but the risk conferred by these common variants is low. Each variant alone does not reach a two-fold increase melanoma risk (89). These genes are involved in different biological processes. There is a group of genes involved in the nevi count and pigmentation including: agouti signaling protein (ASIP) encoding an antagonist of α-MSH, Tyrosinase (TYR) encoding a protein implicated in eye color determination and tanning ability, Tyrosinase-related protein 1 (TYRP1) encoding a protein that stabilizes TYR, Oculocutaneous albinism II (OCA2) playing a role in eye color and pigmentation (90), Methylthioadenosine phosphorylase (MTAP) also implicated in human pigmentation and paired box 3 (PAX3) involved in face and eye development and also associated with nevi count (91). There are also genes involved in the immunologic system such as some interleukins (IL-10, IL-1β), tumor necrosis factor alpha (TNF-α), human leukocyte antigen (HLA) class II genes (92) or interferon regulatory factor 4 (IRF4) (93). Another gene group is related with metabolism: cytochrome P450 family 2 (CYP2D6) that plays a role in the lipid metabolism, genes encoding glutathione transferases (GSTM1, GSTT1 and GSTP1) (94), fat mass and obesity associated (FTO) encoding a protein related with the non-heme iron enzymes (95) and Vitamin D receptor (VDR) involved in mineral metabolism. Poly ADP-ribose polymerase 1 (PARP1), which encodes a chromatin-associated enzyme that modifies nuclear proteins, is also associated with melanoma (93).
Low to moderate risk genes have a weak impact on melanoma susceptibility and families with these variants usually have one or occasionally two melanoma cases. But if a combination of low to moderate variants is inherited, more melanoma cases could be present in the family. However, as UV radiation is the main environmental risk factor for melanoma, families with low to moderate risk variants living in areas with an increased UV radiation could have more melanoma cases (96).
Melanoma genetic counseling and management
Genetic counseling and risk assessment is the process of identifying and counseling individuals at increased risk of developing cancer, and distinguishing between those at high risk (high penetrance genes/families), those at a moderate increased risk (multifactorial etiology or low to moderate penetrance alleles), and those at average risk (97). This information can also be used to assess other at risk individuals within the family. Genetic counseling is offered to melanoma-prone families to better understand the meaning of the disease and genetic susceptibility, the inheritance pattern, the option of genetic testing, the understanding of all the possible results and the primary and secondary prevention of melanoma as well (96). The process includes melanoma risk assessment, the possibility of genetic testing, informed consent, disclosure of test results and psychosocial assessment as in other cancer genetic counseling or assessment (96,97). Doctors should refer patients with a personal and/or family history of melanoma, with features suggestive of having an increased melanoma risk, to a melanoma genetic assessment unit. Genetic counseling often implies genetic testing, but all patients can benefit from genetic counseling, even if they are not candidates for genetic testing. A broad number of reports highlight that genetic counseling in cancer is useful for the screening and management of patients (97). Genetic counseling aims to help patients and families. Genetic counselors should inform patients about the genetic risk factors for melanoma and other cancers, the indications for genetic testing, the chance of detecting a mutation, the possible reports (positive, negative or inconclusive), the possible implications for other family members, provide psychological assessment, and also offer recommendations for cancer screening and UV protection and how these might change with testing (96). Although mutations can be detected in these patients, families and patients negative for a familial mutation still have up to a two-fold greater risk of developing melanoma than that of the general population, due to other melanoma susceptibility genes and environmental factors shared between families (43,98).
To date, in melanoma genetic counseling, genetic testing is mainly focused on CDKN2A and CDK4. However, its use in clinical practice has been controversial due to the variation in the estimates of CDKN2A mutation penetrance (28-91%) depending on the study design, the ethnic background, UV exposure and co-inheritance of low to moderate predisposing variants (such as MC1R variants) (99). It is also difficult to assess patients because individuals negative for the mutation segregating in the family are still at increased risk of melanoma, compared with the general population. Genetic testing for melanoma predisposition mutations can be used in clinical practice under adequate selection criteria and giving a valid test interpretation and genetic counseling to the individual. People should understand that the interpretation of test results is difficult and the potential impact on clinical management is limited. Candidates for genetic testing should only be individuals with at least a 10% chance of carrying a mutation before the test is performed. These individuals should belong to melanoma-prone families or families with melanoma-related cancers (sarcoma, early onset breast cancer, brain tumors or pancreatic cancer) and/or have multiple primary melanomas. Patients with an early age at onset or with multiple or atypical nevi do not fulfill criteria for genetic testing, unless they also have a family history of melanoma (96,100,101). Leachman and colleagues described a very useful rule to select patients for genetic testing in melanoma according to the melanoma incidence in the general population and the prevalence of mutations in each region. In countries with a low melanoma incidence, such as Southern European countries, the selection criteria for genetic counseling should follow the rule of two: individuals with two (synchronous or metachronous) primary melanomas and/or families with at least one invasive melanoma and one or more other diagnoses of melanoma and/or pancreatic cancers among first- or second-degree relatives on the same side of the family. Whilst countries with a moderate to high melanoma incidence, such as the USA and Northern European countries, should follow the rule of three: individuals with three or more primary invasive melanomas and families with at least one invasive melanoma and two more cases of melanoma and/or pancreatic cancer among first- or second-degree relatives on the same side of the family. For very high melanoma incidence countries, such as Australia, the rule of four may be suggested (102).
When genetic testing detects a melanoma predisposing mutation in a family, a screening cascade of individuals at risk is recommended. Individuals carrying CDKN2A mutations are included in prevention and early-detection programs that include the use of sun protection, dermatologic screening and self-skin examination, as an increased skin-cancer screening and surveillance by physicians and self-skin examination results in earlier detection of thinner melanomas (103-105). It has been demonstrated that melanoma genetic counseling has a positive impact on the improvement on total body skin examination and self-skin examination in unaffected individuals carrying germline mutations, after test reporting, while affected carriers maintain high levels of screening adherence (106). Furthermore, after melanoma genetic counseling unaffected members of high risk melanoma families report improvements in daily routine sun protection, showing that genetic counseling may motivate sustained improvements in prevention behaviors (107). Thus it is very important for both melanoma patients and unaffected individuals to be included in genetic counseling programs. Since an increased presence of smoking-related cancers has been detected in CDKN2A mutated families, advice to avoid smoking should also be included in the prevention programs (27,48). To date, individuals carrying CDKN2A mutations should be aware of the current lack of effective screening guidelines for pancreatic cancer (108), although magnetic resonance imaging surveillance can detect early-stage resectable pancreatic tumors (109).
In families where no CDKN2A mutation is identified, it should be stressed that the family is still at increased risk of melanoma on the basis of the family history. No further information on cancer risk is obtained, so these families should be managed according to family history (96). More studies should be necessary to understand the role of the other high-risk genes described to date (BAP1, POT1, ACD, TERF2IP, TERT, CXCL1), which methodology is better to asses them, or the role of the combination of the presence of different low to moderate risk variants, in CDKN2A wild-type families. The inclusion criteria for genetic testing of some of these genes also may differ. For example, BAP1 genetic testing should be offered to families with a broader tumor spectrum including cutaneous and uveal melanoma, mesothelioma, RCC, and also with other skin tumors such as atypical intradermal tumors or basal cell carcinomas. The prevention and early detection programs for patients with mutations in BAP1 should be modified based on the information provided of this cancer syndrome. BAP1 mutation carriers should be closely monitored with, as an example, bi-annual dermatological and annual ophthalmological examinations and pulmonary/renal evaluations by imaging techniques (110). Moreover, many wild-type families for the known genes should carry private mutations that should be assessed by new approaches such as next generation sequencing, in order to detect the mutation responsible for the inherited susceptibility in the family and then improve the genetic counseling specifically in that family.
Conclusions
The new improvements in technology have led to the identification of new genes involved in melanoma susceptibility. However, the knowledge on melanoma susceptibility to date allows us to explain less than 30% of the genetic susceptibility in melanoma-prone families. In some cases the genetic susceptibility may be explained by the accumulation of the presence of low to moderate risk variants in the affected individuals. It is also possible that family aggregation can be explained by shared environmental exposures or also by chance. However next generation techniques can allow the identification of new genes involved in melanoma susceptibility.
Melanoma genetic counseling is important for patients and relatives to better understand the meaning of the disease and the genetic susceptibility. New studies are needed to assess the inclusion in genetic testing of the new genes related to melanoma susceptibility. However, knowing the genetic test results can also be important to refine the genetic counseling in the family as some of the genes involved in melanoma susceptibility also predispose to other tumors. For this reason it is also important to analyze the role that the new genes identified in melanoma may play in the risk to develop other cancers.
Acknowledgements
Funding: The research at the Melanoma Unit in Barcelona is partially funded by Spanish Fondo de Investigaciones Sanitarias grants 09/01393 and 12/00840; CIBER de Enfermedades Raras of the Instituto de Salud Carlos III, Spain; AGAUR 2009 SGR 1337 and AGAUR 2014 SGR 603 of the Catalan Government, Spain; European Commission under the 6th Framework Programme, Contract No.LSHC-CT-2006-018702 (GenoMEL) and by the European Commission under the 7th Framework Programme, Diagnoptics; The National Cancer Institute (NCI) of the US National Institute of Health (NIH) (CA83115) and a grant from “Fundació La Marató de TV3, 201331-30”, Catalonia, Spain. The work was carried out at the Esther Koplowitz Center, Barcelona. Miriam Potrony is the recipient of a PhD Fellowship (PFIS) from Instituto de Salud Carlos III, Spain.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin 2014;64:9-29. [PubMed]
- Lens MB, Dawes M. Global perspectives of contemporary epidemiological trends of cutaneous malignant melanoma. Br J Dermatol 2004;150:179-85. [PubMed]
- Bleyer A, Viny A, Barr R. Cancer in 15- to 29-year-olds by primary site. Oncologist 2006;11:590-601. [PubMed]
- Hanly P, Soerjomataram I, Sharp L. Measuring the societal burden of cancer: the cost of lost productivity due to premature cancer-related mortality in Europe. Int J Cancer 2015;136:E136-45. [PubMed]
- Balch CM, Gershenwald JE, Soong SJ, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol 2009;27:6199-206. [PubMed]
- Gandini S, Sera F, Cattaruzza MS, et al. Meta-analysis of risk factors for cutaneous melanoma: II. Sun exposure. Eur J Cancer 2005;41:45-60. [PubMed]
- Vink AA, Roza L. Biological consequences of cyclobutane pyrimidine dimers. J Photochem Photobiol B 2001;65:101-4. [PubMed]
- Hofmann-Wellenhof R, Soyer HP, Wolf IH, et al. Ultraviolet radiation of melanocytic nevi: a dermoscopic study. Arch Dermatol 1998;134:845-50. [PubMed]
- Stanganelli I, Bauer P, Bucchi L, et al. Critical effects of intense sun exposure on the expression of epiluminescence microscopy features of acquired melanocytic nevi. Arch Dermatol 1997;133:979-82. [PubMed]
- Tronnier M, Wolff HH. UV-irradiated melanocytic nevi simulating melanoma in situ. Am J Dermatopathol 1995;17:1-6. [PubMed]
- Carrera C, Puig-Butille JA, Aguilera P, et al. Impact of sunscreens on preventing UVR-induced effects in nevi: in vivo study comparing protection using a physical barrier vs sunscreen. JAMA Dermatol 2013;149:803-13. [PubMed]
- Green AC, Williams GM, Logan V, et al. Reduced melanoma after regular sunscreen use: randomized trial follow-up. J Clin Oncol 2011;29:257-63. [PubMed]
- Gandini S, Sera F, Cattaruzza MS, et al. Meta-analysis of risk factors for cutaneous melanoma: I. Common and atypical naevi. Eur J Cancer 2005;41:28-44. [PubMed]
- Gandini S, Sera F, Cattaruzza MS, et al. Meta-analysis of risk factors for cutaneous melanoma: III. Family history, actinic damage and phenotypic factors. Eur J Cancer 2005;41:2040-59. [PubMed]
- Levi F, Randimbison L, Te VC, et al. High constant incidence rates of second cutaneous melanomas. Int J Cancer 2005;117:877-9. [PubMed]
- Ferrone CR, Ben Porat L, Panageas KS, et al. Clinicopathological features of and risk factors for multiple primary melanomas. JAMA 2005;294:1647-54. [PubMed]
- Winship IM, Dudding TE. Lessons from the skin--cutaneous features of familial cancer. Lancet Oncol 2008;9:462-72. [PubMed]
- Florell SR, Boucher KM, Garibotti G, et al. Population-based analysis of prognostic factors and survival in familial melanoma. J Clin Oncol 2005;23:7168-77. [PubMed]
- Anderson DE, Badzioch MD. Hereditary cutaneous malignant melanoma: a 20-year family update. Anticancer Res 1991;11:433-7. [PubMed]
- Puig S, Malvehy J, Badenas C, et al. Role of the CDKN2A locus in patients with multiple primary melanomas. J Clin Oncol 2005;23:3043-51. [PubMed]
- Chudnovsky Y, Khavari PA, Adams AE. Melanoma genetics and the development of rational therapeutics. J Clin Invest 2005;115:813-24. [PubMed]
- Lou Z, Chen J. Cellular senescence and DNA repair. Exp Cell Res 2006;312:2641-6. [PubMed]
- Cannon-Albright LA, Goldgar DE, Meyer LJ, et al. Assignment of a locus for familial melanoma, MLM, to chromosome 9p13-p22. Science 1992;258:1148-52. [PubMed]
- Hussussian CJ, Struewing JP, Goldstein AM, et al. Germline p16 mutations in familial melanoma. Nat Genet 1994;8:15-21. [PubMed]
- Aoude LG, Wadt KA, Pritchard AL, et al. Genetics of familial melanoma: 20 years after CDKN2A. Pigment Cell Melanoma Res 2015;28:148-60. [PubMed]
- Goldstein AM, Chan M, Harland M, et al. Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. J Med Genet 2007;44:99-106. [PubMed]
- Potrony M, Puig-Butillé JA, Aguilera P, et al. Increased prevalence of lung, breast, and pancreatic cancers in addition to melanoma risk in families bearing the cyclin-dependent kinase inhibitor 2A mutation: implications for genetic counseling. J Am Acad Dermatol 2014;71:888-95. [PubMed]
- de Torre C, Garcia-Casado Z, Martinez-Escribano JA, et al. Influence of loss of function MC1R variants in genetic susceptibility of familial melanoma in Spain. Melanoma Res 2010;20:342-8. [PubMed]
- Pedace L, De Simone P, Castori M, et al. Clinical features predicting identification of CDKN2A mutations in Italian patients with familial cutaneous melanoma. Cancer Epidemiol 2011;35:e116-20. [PubMed]
- Casula M, Muggiano A, Cossu A, et al. Role of key-regulator genes in melanoma susceptibility and pathogenesis among patients from South Italy. BMC Cancer 2009;9:352. [PubMed]
- Bruno W, Ghiorzo P, Battistuzzi L, et al. Clinical genetic testing for familial melanoma in Italy: a cooperative study. J Am Acad Dermatol 2009;61:775-82. [PubMed]
- Nikolaou V, Kang X, Stratigos A, et al. Comprehensive mutational analysis of CDKN2A and CDK4 in Greek patients with cutaneous melanoma. Br J Dermatol 2011;165:1219-22. [PubMed]
- Veinalde R, Ozola A, Azarjana K, et al. Analysis of Latvian familial melanoma patients shows novel variants in the noncoding regions of CDKN2A and that the CDK4 mutation R24H is a founder mutation. Melanoma Res 2013;23:221-6. [PubMed]
- Lukowsky A, Schafer-Hesterberg G, Sterry W, et al. Germline CDKN2A/p16 mutations are rare in multiple primary and familial malignant melanoma in German patients. J Dermatol Sci 2008;49:163-5. [PubMed]
- Wadt KA, Aoude LG, Krogh L, et al. Molecular characterization of melanoma cases in Denmark suspected of genetic predisposition. PLoS One 2015;10:e0122662. [PubMed]
- Helgadottir H, Höiom V, Tuominen R, et al. CDKN2a mutation-negative melanoma families have increased risk exclusively for skin cancers but not for other malignancies. Int J Cancer 2015;137:2220-6. [PubMed]
- Larre Borges A, Cuéllar F, Puig-Butillé JA, et al. CDKN2A mutations in melanoma families from Uruguay. Br J Dermatol 2009;161:536-41. [PubMed]
- de Ávila AL, Krepischi AC, Moredo LF, et al. Germline CDKN2A mutations in Brazilian patients of hereditary cutaneous melanoma. Fam Cancer 2014;13:645-9. [PubMed]
- Grazziotin TC, Rey MC, Bica CG, et al. Genetic variations of patients with familial or multiple melanoma in Southern Brazil. J Eur Acad Dermatol Venereol 2013;27:e179-85. [PubMed]
- Goldstein AM, Chan M, Harland M, et al. High-risk melanoma susceptibility genes and pancreatic cancer, neural system tumors, and uveal melanoma across GenoMEL. Cancer Res 2006;66:9818-28. [PubMed]
- Auroy S, Avril MF, Chompret A, et al. Sporadic multiple primary melanoma cases: CDKN2A germline mutations with a founder effect. Genes Chromosomes Cancer 2001;32:195-202. [PubMed]
- Harland M, Cust AE, Badenas C, et al. Prevalence and predictors of germline CDKN2A mutations for melanoma cases from Australia, Spain and the United Kingdom. Hered Cancer Clin Pract 2014;12:20. [PubMed]
- Bishop DT, Demenais F, Goldstein AM, et al. Geographical variation in the penetrance of CDKN2A mutations for melanoma. J Natl Cancer Inst 2002;94:894-903. [PubMed]
- Vasen HF, Gruis NA, Frants RR, et al. Risk of developing pancreatic cancer in families with familial atypical multiple mole melanoma associated with a specific 19 deletion of p16 (p16-Leiden). Int J Cancer 2000;87:809-11. [PubMed]
- Mukherjee B, Delancey JO, Raskin L, et al. Risk of non-melanoma cancers in first-degree relatives of CDKN2A mutation carriers. J Natl Cancer Inst 2012;104:953-6. [PubMed]
- Ghiorzo P, Fornarini G, Sciallero S, et al. CDKN2A is the main susceptibility gene in Italian pancreatic cancer families. J Med Genet 2012;49:164-70. [PubMed]
- Borg A, Sandberg T, Nilsson K, et al. High frequency of multiple melanomas and breast and pancreas carcinomas in CDKN2A mutation-positive melanoma families. J Natl Cancer Inst 2000;92:1260-6. [PubMed]
- Helgadottir H, Hoiom V, Jonsson G, et al. High risk of tobacco-related cancers in CDKN2A mutation-positive melanoma families. J Med Genet 2014;51:545-52. [PubMed]
- Zuo L, Weger J, Yang Q, et al. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat Genet 1996;12:97-9. [PubMed]
- Puntervoll HE, Yang XR, Vetti HH, et al. Melanoma prone families with CDK4 germline mutation: phenotypic profile and associations with MC1R variants. J Med Genet 2013;50:264-70. [PubMed]
- Aoude LG, Wadt K, Bojesen A, et al. A BAP1 mutation in a Danish family predisposes to uveal melanoma and other cancers. PLoS One 2013;8:e72144. [PubMed]
- Abdel-Rahman MH, Pilarski R, Cebulla CM, et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet 2011;48:856-9. [PubMed]
- Carbone M, Ferris LK, Baumann F, et al. BAP1 cancer syndrome: malignant mesothelioma, uveal and cutaneous melanoma, and MBAITs. J Transl Med 2012;10:179. [PubMed]
- Carbone M, Yang H, Pass HI, et al. BAP1 and cancer. Nat Rev Cancer 2013;13:153-9. [PubMed]
- Njauw CN, Kim I, Piris A, et al. Germline BAP1 inactivation is preferentially associated with metastatic ocular melanoma and cutaneous-ocular melanoma families. PLoS One 2012;7:e35295. [PubMed]
- Popova T, Hebert L, Jacquemin V, et al. Germline BAP1 Mutations Predispose to Renal Cell Carcinomas. Am J Hum Genet 2013;92:974-80. [PubMed]
- Wiesner T, Obenauf AC, Murali R, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet 2011;43:1018-21. [PubMed]
- Wadt KA, Aoude LG, Johansson P, et al. A recurrent germline BAP1 mutation and extension of the BAP1 tumor predisposition spectrum to include basal cell carcinoma. Clin Genet 2015;88:267-72. [PubMed]
- de la Fouchardière A, Cabaret O, Savin L, et al. Germline BAP1 mutations predispose also to multiple basal cell carcinomas. Clin Genet 2015;88:273-7. [PubMed]
- Höiom V, Edsgärd D, Helgadottir H, et al. Hereditary uveal melanoma: a report of a germline mutation in BAP1. Genes Chromosomes Cancer 2013;52:378-84. [PubMed]
- Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 2010;330:1410-3. [PubMed]
- Maerker DA, Zeschnigk M, Nelles J, et al. BAP1 germline mutation in two first grade family members with uveal melanoma. Br J Ophthalmol 2014;98:224-7. [PubMed]
- Pilarski R, Cebulla CM, Massengill JB, et al. Expanding the clinical phenotype of hereditary BAP1 cancer predisposition syndrome, reporting three new cases. Genes Chromosomes Cancer 2014;53:177-82. [PubMed]
- Farley MN, Schmidt LS, Mester JL, et al. A novel germline mutation in BAP1 predisposes to familial clear-cell renal cell carcinoma. Mol Cancer Res 2013;11:1061-71. [PubMed]
- Wadt K, Choi J, Chung JY, et al. A cryptic BAP1 splice mutation in a family with uveal and cutaneous melanoma, and paraganglioma. Pigment Cell Melanoma Res 2012;25:815-8. [PubMed]
- Yang XR, Brown K, Landi MT, et al. Duplication of CXC chemokine genes on chromosome 4q13 in a melanoma-prone family. Pigment Cell Melanoma Res 2012;25:243-7. [PubMed]
- O'Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol 2010;11:171-81. [PubMed]
- Valdes AM, Andrew T, Gardner JP, et al. Obesity, cigarette smoking, and telomere length in women. Lancet 2005;366:662-4. [PubMed]
- Han J, Qureshi AA, Prescott J, et al. A prospective study of telomere length and the risk of skin cancer. J Invest Dermatol 2009;129:415-21. [PubMed]
- Iles MM, Bishop DT, Taylor JC, et al. The effect on melanoma risk of genes previously associated with telomere length. J Natl Cancer Inst 2014.106. [PubMed]
- Horn S, Figl A, Rachakonda PS, et al. TERT promoter mutations in familial and sporadic melanoma. Science 2013;339:959-61. [PubMed]
- Robles-Espinoza CD, Harland M, Ramsay AJ, et al. POT1 loss-of-function variants predispose to familial melanoma. Nat Genet 2014;46:478-81. [PubMed]
- Shi J, Yang XR, Ballew B, et al. Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat Genet 2014;46:482-6. [PubMed]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet 2008;42:301-34. [PubMed]
- Aoude LG, Pritchard AL, Robles-Espinoza CD, et al. Nonsense mutations in the shelterin complex genes ACD and TERF2IP in familial melanoma. J Natl Cancer Inst 2014.107. [PubMed]
- Dessinioti C, Antoniou C, Katsambas A, et al. Melanocortin 1 receptor variants: functional role and pigmentary associations. Photochem Photobiol 2011;87:978-87. [PubMed]
- Williams PF, Olsen CM, Hayward NK, et al. Melanocortin 1 receptor and risk of cutaneous melanoma: a meta-analysis and estimates of population burden. Int J Cancer 2011;129:1730-40. [PubMed]
- Flanagan N, Healy E, Ray A, et al. Pleiotropic effects of the melanocortin 1 receptor (MC1R) gene on human pigmentation. Hum Mol Genet 2000;9:2531-7. [PubMed]
- Raimondi S, Sera F, Gandini S, et al. MC1R variants, melanoma and red hair color phenotype: a meta-analysis. Int J Cancer 2008;122:2753-60. [PubMed]
- Fargnoli MC, Gandini S, Peris K, et al. MC1R variants increase melanoma risk in families with CDKN2A mutations: a meta-analysis. Eur J Cancer 2010;46:1413-20. [PubMed]
- Córdoba-Lanús E, Hernández-Jiménez JG, Medina-Coello C, et al. MC1R gene variants and sporadic malignant melanoma susceptibility in the Canary Islands population. Arch Dermatol Res 2014;306:51-8. [PubMed]
- Puig-Butillé JA, Carrera C, Kumar R, et al. Distribution of MC1R variants among melanoma subtypes: p.R163Q is associated with lentigo maligna melanoma in a Mediterranean population. Br J Dermatol 2013;169:804-11. [PubMed]
- Yokoyama S, Woods SL, Boyle GM, et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature 2011;480:99-103. [PubMed]
- Bertolotto C, Lesueur F, Giuliano S, et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature 2011;480:94-8. [PubMed]
- Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med 2006;12:406-14. [PubMed]
- Ghiorzo P, Pastorino L, Queirolo P, et al. Prevalence of the E318K MITF germline mutation in Italian melanoma patients: associations with histological subtypes and family cancer history. Pigment Cell Melanoma Res 2013;26:259-62. [PubMed]
- Sturm RA, Fox C, McClenahan P, et al. Phenotypic characterization of nevus and tumor patterns in MITF E318K mutation carrier melanoma patients. J Invest Dermatol 2014;134:141-9. [PubMed]
- Berwick M, MacArthur J, Orlow I, et al. MITF E318K's effect on melanoma risk independent of, but modified by, other risk factors. Pigment Cell Melanoma Res 2014;27:485-8. [PubMed]
- Ward KA, Lazovich D, Hordinsky MK. Germline melanoma susceptibility and prognostic genes: a review of the literature. J Am Acad Dermatol 2012;67:1055-67. [PubMed]
- Fernandez LP, Milne RL, Pita G, et al. Pigmentation-related genes and their implication in malignant melanoma susceptibility. Exp Dermatol 2009;18:634-42. [PubMed]
- Ogbah Z, Badenas C, Harland M, et al. Evaluation of PAX3 genetic variants and nevus number. Pigment Cell Melanoma Res 2013;26:666-76. [PubMed]
- Planelles D, Nagore E, Moret A, et al. HLA class II polymorphisms in Spanish melanoma patients: homozygosity for HLA-DQA1 locus can be a potential melanoma risk factor. Br J Dermatol 2006;154:261-6. [PubMed]
- Peña-Chilet M, Blanquer-Maceiras M, Ibarrola-Villava M, et al. Genetic variants in PARP1 (rs3219090) and IRF4 (rs12203592) genes associated with melanoma susceptibility in a Spanish population. BMC Cancer 2013;13:160. [PubMed]
- Ibarrola-Villava M, Martin-Gonzalez M, Lazaro P, et al. Role of glutathione S-transferases in melanoma susceptibility: association with GSTP1 rs1695 polymorphism. Br J Dermatol 2012;166:1176-83. [PubMed]
- Iles MM, Law MH, Stacey SN, et al. A variant in FTO shows association with melanoma risk not due to BMI. Nat Genet 2013;45:428-32, 432e1.
- Badenas C, Aguilera P, Puig-Butille JA, et al. Genetic counseling in melanoma. Dermatol Ther 2012;25:397-402. [PubMed]
- Riley BD, Culver JO, Skrzynia C, et al. Essential elements of genetic cancer risk assessment, counseling, and testing: updated recommendations of the National Society of Genetic Counselors. J Genet Couns 2012;21:151-61. [PubMed]
- Udayakumar D, Tsao H. Melanoma genetics: an update on risk-associated genes. Hematol Oncol Clin North Am 2009;23:415-29. vii. [PubMed]
- Aspinwall LG, Leaf SL, Dola ER, et al. CDKN2A/p16 genetic test reporting improves early detection intentions and practices in high-risk melanoma families. Cancer Epidemiol Biomarkers Prev 2008;17:1510-9. [PubMed]
- Hansen CB, Wadge LM, Lowstuter K, et al. Clinical germline genetic testing for melanoma. Lancet Oncol 2004;5:314-9. [PubMed]
- Tsao H, Zhang X, Kwitkiwski K, et al. Low prevalence of germline CDKN2A and CDK4 mutations in patients with early-onset melanoma. Arch Dermatol 2000;136:1118-22. [PubMed]
- Leachman SA, Carucci J, Kohlmann W, et al. Selection criteria for genetic assessment of patients with familial melanoma. J Am Acad Dermatol 2009;61:677.e1-14.
- Carli P, De Giorgi V, Palli D, et al. Dermatologist detection and skin self-examination are associated with thinner melanomas: results from a survey of the Italian Multidisciplinary Group on Melanoma. Arch Dermatol 2003;139:607-12. [PubMed]
- Salerni G, Carrera C, Lovatto L, et al. Benefits of total body photography and digital dermatoscopy ("two-step method of digital follow-up") in the early diagnosis of melanoma in patients at high risk for melanoma. J Am Acad Dermatol 2012;67:e17-27. [PubMed]
- Salerni G, Lovatto L, Carrera C, et al. Melanomas detected in a follow-up program compared with melanomas referred to a melanoma unit. Arch Dermatol 2011;147:549-55. [PubMed]
- Aspinwall LG, Taber JM, Leaf SL, et al. Melanoma genetic counseling and test reporting improve screening adherence among unaffected carriers 2 years later. Cancer Epidemiol Biomarkers Prev 2013;22:1687-97. [PubMed]
- Aspinwall LG, Taber JM, Kohlmann W, et al. Unaffected family members report improvements in daily routine sun protection 2 years following melanoma genetic testing. Genet Med 2014;16:846-53. [PubMed]
- de Snoo FA, Bishop DT, Bergman W, et al. Increased risk of cancer other than melanoma in CDKN2A founder mutation (p16-Leiden)-positive melanoma families. Clin Cancer Res 2008;14:7151-7. [PubMed]
- Vasen HF, Wasser M, van Mil A, et al. Magnetic resonance imaging surveillance detects early-stage pancreatic cancer in carriers of a p16-Leiden mutation. Gastroenterology 2011;140:850-6. [PubMed]
- Battaglia A. The Importance of Multidisciplinary Approach in Early Detection of BAP1 Tumor Predisposition Syndrome: Clinical Management and Risk Assessment. Clin Med Insights Oncol 2014;8:37-47. [PubMed]


