
TNF-α-stimulated nucleus pulposus cells induce cell apoptosis through the release of exosomal miR-16 targeting IGF-1 and IGF-1R in rats
Introduction
Intervertebral disc herniation (IDH) is a major cause of low back pain (LBP), which is a leading contributor to years lived with a disability worldwide (1). The nucleus pulposus (NP), which consists of NP cells (NPCs) and abundant extracellular matrix, is located at the center of the intervertebral disc and bears most of the axial compression pressure of the spine. Disc samples and clinical images from IDH patients are commonly characterized by damage to the annulus fibrosus (AF), the outer layer of the disc, and the herniation of NP tissue inside the disc. Consistent with the low signal intensity observed on T2-weighted magnetic resonance imaging (MRI), immunohistochemical staining of IDH NP samples typically reveals a lack of collagens and proteoglycans (PGs), which are major components of the NP matrix, and decreasing numbers of NPCs (2).
Apoptosis, also known as programmed cell death (PCD), is a programmed and orderly cell death process induced to maintain cellular homeostasis. Previous studies have reported high rates of NPC apoptosis in vitro and the high level of apoptotic markers in NP samples from both IDH patients and animal models (3,4). Some proinflammatory factors, such as nitric oxide, tumor necrosis factor-α (TNF-α), and interleukin-1β (IL-1β), can induce cell apoptosis via various pathways. For example, TNF-α can bind to TNF receptor 1 (TNFR1), a membrane receptor that triggers a downstream caspase activation cascade, inducing apoptosis in NPCs (5). Therefore, TNF-α may serve as a key initiating factor in disc degeneration (6).
Insulin-like growth factor-1 (IGF-1) is a natural polypeptide hormone that plays an important role in growth stimulation during childhood and is involved in tissue regeneration in adults (7). Previous studies have demonstrated that IGF-1 is critical to the maintenance of disc homeostasis, promoting both cell proliferation and the synthesis of extracellular matrix in a dose-dependent manner (8,9). The major functional receptor for IGF-I, IGF-1R, is a receptor tyrosine kinase that can activate the phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt) signaling pathway (10,11) and then serves as a natural stimulator of cell growth and proliferation and a potent inhibitor of apoptosis in NPCs (12).
Exosomes are a group of membranous, nano-sized particles originating from endosomes which contain bioactive lipids, proteins, and nucleic acids. One group of important exosomal cargoes, known as microRNAs (miRNAs), are small, non-coding RNAs that function as negative gene expression regulators. Many studies have reported the protective role played by exosomal miRNAs that are derived from the cells involved in disc degeneration diseases, such as IDH (13-16). Recently, we reported that the exosomal miR-27a derived from autophagy-activated healthy NPCs could repress IL-1β-induced NP matrix degradation by targeting matrix metalloprotease 13 (MMP-13) (17).
However, whether exosomes released from NPCs under pathological conditions, such as inflammation, can affect the proliferation and apoptosis of normal NPCs remains largely unknown. In this study, we investigated the effects of exosomes derived from TNF-α-stimulated NPCs on the induction of apoptosis in normal NPCs and the relationship between IGF-1 expression and exposure to these exosomes. This study may help us understand the relationship between inflammation and apoptosis in the pathology of intervertebral disc degeneration.
We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-227).
Methods
Rat NPC culture and identification
The animal experiments performed in this study were approved by the Animal Care and Use Committee of Fudan University (Shanghai, China, No. Y2021-228). Twenty-four 2-month-old male Sprague-Dawley (SD) rats were obtained from the Department of Laboratory Animal Science. The rats were euthanized with an overdose of 10% (w/v) chloral hydrate (3.5 mL/kg body weight). The lumbar spinal columns were removed en bloc. The NP tissue was separated from the lumbar disc, without the outer AF layer and cartilaginous endplates though microsurgery, followed by digestion with 0.1% collagenase II for 4 h at 37 °C. The tissues were propagated in a T25 culture flask, without changing the media, until the NPCs began to grow out of the tissue. The NPCs were seeded at the density of 3×104 cells/mL in Dulbecco’s modified Eagle’s medium (DMEM; Hyclone, Thermo Scientific)-high glucose supplemented with 10% fetal bovine serum (FBS; Hyclone, Thermo Scientific) and antibiotics (1% penicillin/streptomycin). All procedures were aseptically performed on a clean laboratory bench. The second passage of NPCs was used for the following experiments. All NPCs were maintained in a hypoxia incubator maintained at 1% O2 and 5% CO2 at 37 °C. Upon reaching 70–80% cell confluence, the cells were harvested using 0.25% trypsin-ethylenediaminetetraacetic acid (EDTA; Invitrogen) for further experiments.
Cell density was determined by bright-field cell counter following the instruments (DeNovix, USA) of manufactures. All animal experiments were carried out in accordance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals.
Alcian blue and toluidine blue staining, and collagen II immunofluorescence for the identification of rat NPCs
Cells were seeded at the density of 3×104 cells/mL and cultured in 6-well plate. Upon reaching 70–80% cell confluence, NP cells were fixed with 4% paraformaldehyde for 15 min, and washed 3 times with phosphate-buffered saline (PBS). Then, 1% Alcian blue dye (prepared with acetic acid) or 1% toluidine blue dye (prepared with 70% alcohol) was added to the wells and incubated at room temperature for 60 min. After an additional 3 washes with PBS, the NPCs were observed under an inverted microscope.
For immunofluorescence staining, fixed NPCs were sequentially incubated with the primary antibodies of collagen II (#ab34712, 1:200; Abcam) and secondary antibodies (#ab150078, 1:400; Abcam). The samples were then washed with PBS, mounted in glycerol gel which contained DAPI for nuclei staining (Beyotime, China), and observed under an FV3000 confocal laser scanning microscope (Olympus, Tokyo, Japan).
Cell viability test
Briefly, NPCs (density: 3×103 cells per well) were seeded in a 96-well plate. The cell viability was examined by using a Cell Counting Kit-8 (CCK-8; Beyotime, China) following the manufacturer’s protocols. The NPCs were treated with different concentration of TNF-α for 36 h. Optical density (OD) was measured at a wavelength of 450 nm using a Gen5 microplate reader (BioTek Instruments, Inc., Winooski, VT, USA).
Exosome isolation and identification
NPCs were seeded at 3×106 cells per T75 culture flask and cultured in 15 mL of DMEM-high glucose medium with 10% exosome-depleted FBS. The exosome-depleted FBS was obtained by ultracentrifugation, as described below. The medium was collected after 36 h of 10 ng/mL TNF-α pretreatment, followed by gradient centrifugation to remove debris, and ultracentrifugation at 120,000 ×g for 120 min, as previously described (17). The pellet was resuspended in PBS, quantified by bicinchoninic acid (BCA) protein assay (Beyotime, China), and cryopreserved at −80 °C.
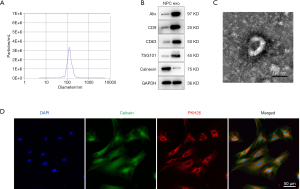
Exosome morphologies were detected using transmission electron microscopy (TEM). Briefly, fresh exosome samples were loaded onto glow-discharged, carbon-coated, copper grids and stained with 1% (w/v) uranyl acetate for 1 min. The stained exosome samples were examined using an H-7650 transmission electron microscope (Hitachi, Japan) at an acceleration voltage of 80 kV.
The sizes and concentrations of the exosomes were assessed using nanoparticle tracking analysis (NTA) instruments, according to the manufacturer’s instructions, and the data were analyzed by ZetaView 8.04.02 SP2 software.
Antibodies for the exosomal markers CD9 (#ab92726, 1:2,000; Abcam), CD63 (#ab134045, 1:2,000; Abcam), apoptosis-linked gene 2-interacting protein X (ALIX, #ab117600, 1:2,000; Abcam), and tumor susceptibility gene 101 (TSG101, #ab30871, 1:1,000; Abcam) and the negative marker calnexin (#ab22595, 1:2,000; Abcam), an endoplasmic reticulum marker protein, were analyzed by Western blot. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH, #ab9485, 1:2,000; Abcam) was used as the loading control.
Exosome uptake by NPCs
Exosomes were labeled with PKH26 (Sigma-Aldrich, USA), according to the manufacturer’s instructions. After ultracentrifugation, as described above, the labeled exosomes were suspended in culture medium and incubated with NPCs for 4 h at 37 °C. Before observation, NPCs were washed twice with PBS. Calcein and Hoechst 33342 (Beyotime, China) were added to stain the cellular components and nuclei of live NPCs for 5 and 2 min, respectively. The samples were then observed under an FV3000 confocal laser scanning microscope (Olympus, Tokyo, Japan).
Western blotting
Cells or exosomes were lysed in radio immunoprecipitation assay (RIPA) buffer (Beyotime, China) mixed with 1× protease inhibitor cocktail (Yeasen, China). Lysates were boiled in 1× sodium dodecyl sulfate (SDS) buffer for 10 min to denature proteins. Proteins were separated via SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to a polyvinylidene fluoride membrane. The membrane was blocked, washed 3 times with Tris-buffered saline (TBS) containing 0.1% Tween-20 (TBST), and incubated with the specific primary antibody overnight at 4 °C. After 3 washes, horseradish peroxidase-labeled secondary antibodies (#ab205718, 1:5,000; Abcam) were added and incubated for one hour at room temperature. After 3 washes with PBS, the protein bands were visualized with enhanced chemiluminescence (ECL) reagent (Share-Bio, China) and visualized using the ChemiScope system (Clinx, China). Semi-quantification of the bands was performed using ImageJ software to measure grey density.
The primary antibodies used are as follows: IGF-1 (#abs119614, 1:1,000; Absin), IGF-1R (#ab 182408, 1:1,000; Abcam), PI3K (#4249, 1:1,000; CST), p-PI3K (#4228, 1:1,000; CST), Bax (#ab32503, 1:1,000; Abcam), Bcl-2 (#ab182858, 1:1,000; Abcam), cleaved caspase-3 (#ab32042, 1:1,000; Abcam), caspase-3 (#ab13847, 1:1,000; Abcam), Akt (#ab38449, 1:1,000; Abcam), and p-Akt (#ab8805, 1:1,000; Abcam). The antibodies for β-actin (#ab8227, 1:2,000; Abcam) and GAPDH (#ab9485, 1:2,000; Abcam) were used as loading controls.
Flow cytometry analysis
NPCs were plated into 6-well plates at a density of 1×105 cells per well. To test the apoptosis-inducing effect of exosomes from TNF-α-treated NPCs on cells, NPCs were treated with 100 mg/mL exosomes for 12 hours. Following treatment, NPC apoptosis rates were evaluated by flow cytometry using an Annexin V/PI apoptosis detection kit (BD Biosciences, USA). NPCs were washed twice with PBS, resuspended in binding buffer and incubated with 5 µL FITC-Annexin V and 5 µL PI for 20 min at room temperature. Staining cells were analysed using the FACScan flow cytometry system (Becton Dickinson, USA).
Dual-luciferase reporter assay
The rat 3'-untranslated region (UTR) of the IGF-1 and IGF-1R genes were amplified by polymerase chain reaction (PCR) using specific primers.
The following primers were used: IGF-1R: forward 5'-TCCTTGGATCCTGAATCTGT-3', reverse 5'-ACGTTGCCTTAGCTTCAGCC-3'; IGF-1: forward 5'- GAGGAGCCTCCCGAGGAACA-3', reverse 5'-CCTAATTTTGTCCTTTTGGG-3'.
Wild-type or mutant IGF-1 and IGF-1R 3'-UTR fragments were inserted into the XbaI restriction sites of the pmirGLO luciferase vector (Promega, USA). For transfection, 293T cells were plated in 96-well plates at 1.2×105 cells per well and co-transfected with the IGF-1 or IGF-1R 3'UTR reporter plasmid and either miR-16 mimics or negative control (NC) using Opti-MEM (Sigma-Aldrich, USA) and Lipofectamine 3000 transfection reagent (Sigma-Aldrich, USA). Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, USA) 36 h after transfection. The optical density of the resulting solution was measured using the EPOCH2T multiplate reader (BioTek, USA).
Transfection of miR-16 inhibitors and mimics
miR-16 inhibitors and mimics were purchased from RiboBIO (China). Both inhibitors and mimics were transfected using the riboFECT CP Reagent (RiboBIO, China), according to the manufacturer’s protocol, at a final concentration of 120 nM. Cells were either harvested or subjected to additional treatments after 36 h transfection.
RNA extraction and quantification
TRIzol reagent (Takara, Japan) was used to extract total RNA. miR-452-5p, miR-190a-5p, miR-301a-3p, and miR-16-5p were reverse-transcribed and quantified using a Bulge-Loop miRNA RT-PCR Quantitation kit (RiboBIO, China). Quantitative real-time-PCR (qRT-PCR) was performed using the QuantStudio5 Real-Time PCR System (ABI, USA). Fold changes in expression were calculated using the comparative threshold cycle (CT) method, using the formula 2−ΔΔCT. U6 and miR-39 (RiboBIO, China) were used as controls for miRNA in cells and exosomes, respectively.
Human NP tissues
Seven NP samples from disc extrusion or sequestration patients who underwent spine fusion were supplied by the Zhongshan Hospital of Fudan University from November 2018 to January 2021. Study approval was granted by the Research Ethics Committee of Zhongshan Hospital (No. Y2021-228). The use of tissues in this study followed the ethical guidelines of the Declaration of Helsinki (as revised in 2013), and each patient provided written informed consent for their tissues to be used in the scientific research. The sex, age and Pfirrmann grade based on MRI characteristics were recorded (Table S1).
Immunohistochemistry
Sections of formalin-fixed, paraffin-embedded NP tissues were subjected to xylene deparaffinization and rehydration with gradient concentrations of ethanol. Then, overnight incubation of the tissue sections was conducted in the presence of IGF-1 (#ab106836, 1:200; Abcam), IGF-1R (#ab 182408, 1:200; Abcam), Bax (#ab32503, 1:200; Abcam), Bcl-2 (#ab182858, 1:200; Abcam), caspase-3 (#ab13847, 1:100; Abcam) and TNF-α (#ab270264, 1:100; Abcam) primary antibodies. The corresponding IgG HRP-conjugated secondary antibodies (#ab205718, 1:400; Abcam) were incubated at room temperature for 1 h. The images were observed with an Olympus DX51 microscope (Olympus, Japan). Data analysis was performed using ImageJ software.
Statistical analysis
All results are displayed as the mean ± standard deviation (SD). All biological experiments were independently repeated three times. Unpaired Student’s t-tests were used to compare two groups. One-way and two-way analyses of variance (ANOVAs) were applied to compare multiple groups. Normality test and the nonparametric tests were also used if the normality assumption was not fulfilled. Data analyses and visualization were performed using GraphPad Prism 7.0. Significance was set at P<0.05.
Results
Identification of NPC-derived exosomes and uptake of exosomes by NPCs
Primary cultured SD rat NPCs were identified by alcian blue and toluidine blue staining and collagen II immunofluorescence. NPCs stained with Alcian blue appeared light blue in coloration (Figure S1A) due to the high expression of aggrecan. Notochord cells existed in a clustered form, containing a large number of vacuoles, and exhibited metachromasia when stained with toluidine blue (Figure S1B,S1C). NPC cytoplasmic compartments contained large amounts of collagen II, confirming that NPCs exhibit cartilage-like characteristics (Figure S1D).
The NTA results revealed that the average exosome diameter from TNF-α-pretreated NPCs was 132.1±37.3 nm (Figure 1A). Exosomes from both normal NPCs and TNF-α-pretreated NPCs expressed the exosomal marker proteins ALIX, CD9, CD63, and TSG101. Calnexin was detected as a negative control, which is uniquely expressed and localized in the endoplasmic reticulum (Figure 1B). TEM revealed that the exosome particles were cup-shaped vesicles with a double-layer membrane structure (Figure 1C). After incubation with NPCs for 4 h, the PKH26-labelled exosomes appeared as red fluorescence in the NPC cytoplasm (Figure 1D), indicating successful exosome uptake by NPCs.
The effects of TNF-α on NPC cell viability and exosome release
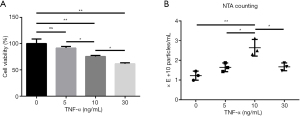
Different doses of TNF-α were used to treat NPCs, and proliferation and cell viability were measured. After 36 h treatment, the cell viability of the 10 ng/mL TNF-α treatment group was 75.5%±1.9%, whereas the cell viability of the 30 ng/mL TNF-α treatment group was only 61.7%±1.7%. The cell viability of the 5 ng/mL TNF-α treatment group was not significantly different from that of the control group (Figure 2A). We also analyzed whether TNF-α could stimulate exosome release by NPCs. As shown in Figure 2B, the NTA data indicated that NPCs treated with 10 ng/mL TNF-α for 36 h released an average of (2.64±0.43)×1010 particles/mL, compared with (1.65±0.22)×1010 particles/mL released by the 5 ng/mL group and (1.67±0.20)×1010 particles/mL released by the 30 ng/mL group. Based on both cell viability and exosome production results, we chose to use the 10 ng/mL TNF-α treatment for all subsequent NPC exosome release experiments.
Exosomes derived from TNF-α treated NPCs induce the apoptosis of normal NPCs and repress IGF-1 expression
We then investigated the biological effects of exosomes derived from TNF-α-pretreated NPCs (TNF-Exos) on normal NPCs. As shown in Figure 3A, incubation with TNF-Exos induced NPC apoptosis, as assessed by flow cytometry. Western blot analysis indicated that TNF-Exos significantly induced the increased expression of the apoptosis marker Bax, increased the cleaved caspase-3/caspase-3 ratio, and reduced the level of the anti-apoptosis marker Bcl-2 (Figure 3B).
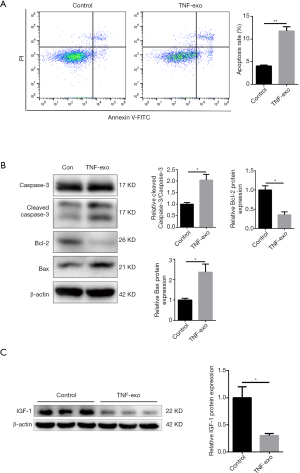
We examined whether the IGF-1 signaling pathway was involved in the TNF-Exo-induced apoptosis of NPCs. Western blot analysis indicated that the IGF-1 expression was significantly reduced by incubation with TNF-Exos (Figure 3C).
miR-16 is abundant in exosomes from TNF-α-pretreated NPCs and regulates both IGF-1 and IGF-1R in normal NPCs
We investigated whether TNF-Exos induced NPC apoptosis and inhibited the IGF-1 signaling pathway through the delivery of exosomal miRNAs.
TargetScan and miRDB are widely used bioinformatics tools for miRNA target prediction, containing miRNA information from 10 species, including humans, rats, and mice (18,19). Indexes, including context score and PCT, are provided to predict target interactions and evolutionary conservation during the selection of potential miRNA-mRNA pairs. We downloaded the predicted rat IGF-1-regulating miRNAs from both tools and extracted the overlapping list, which included 12 miRNAs predicted to bind to the IGF-1 3'UTR (Figure 4A). We ranked the miRNAs according to the prediction scores provided by both platforms and examined the top 4 miRNAs on the list: miR-452-5p, miR-190a-5p, miR-301a-3p, and miR-16-5p. Both exosomes and their parent cells were analyzed.
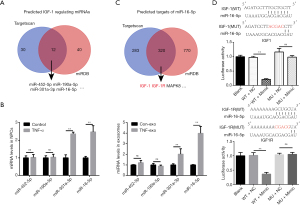
Only miR-301a-3p and miR-16-5p were significantly abundant in both 10 ng/mL TNF-α-pretreated NPCs and TNF-Exos. Interestingly, miR-16-5p had a relatively higher expression level in TNF-Exos than in TNF-α-pretreated NPCs, with fold-changes of 3.99±0.64 in exosomes and 2.47±0.35 in cells (Figure 4B). Therefore, we chose miR-16-5p for further study. We searched for potential miR-16-5p targets in both TargetScan and miRDB to determine the potential downstream biological functions affected by this miRNA. Coincidentally, we found that IGF-1R, the IGF-1 receptor, was also a predicted target of miR-16-5p, with high prediction scores according to both tools (context++, score percentile 89 in TargetScan; target score 82 in miRDB, Full mark: 100; Figure 4C). To verify that the miR-16 binding sites in the 3'UTRs of IGF-1 and IGF-1R mRNA were functional, mutant 3'UTR sequences for the IGF-1 and IGF-1R genes were transfected into luciferase reporter plasmids. As expected, miR-16 mimics exclusively decreased the luciferase activity of the wild-type reporter plasmids for both IGF-1 and IGF-1R, whereas no suppression of activity was observed for the mutant plasmids (Figure 4D).
Exosomes from miR-16 mimic-transfected NPCs repressed IGF-1 and IGF-1R expression and the downstream PI3K/Akt pathway in normal NPCs
Next, we validated the role played by miR-16 in the exosome-induced apoptosis of NPCs. NPCs were transfected with miR-16 mimics or normal control (NC) miRNA (miR-NC). To investigate the contribution of miR-16 to the pro-apoptotic effect of NPC-produced exosomes, we developed miR-16-overexpressing exosomes by transfecting NPCs with miR-16 mimics, followed by exosome isolation. The successful overexpression of miR-16 in both NPCs and exosomes was confirmed by qRT-PCR (Figure 5A).
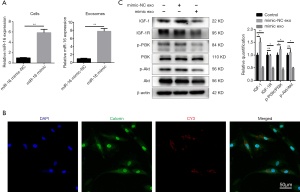
To detect the successful uptake of exosomal miR-16, NPCs were transfected with Cy3-labeled miR-16 mimics, followed by exosome isolation. NPCs incubated with these exosomes exhibited a granular fluorescent pattern within the cytoplasm (Figure 5B). Then, we investigated whether exosomal miR-16 could induce NPC apoptosis through the downregulation of IGF-1/IGF-1R signaling and the subsequent inhibition of the PI3K/Akt pathway.
The incubation of NPCs with exosomes derived from miR-16 mimic-transfected-NPCs resulted in the significant downregulation of both IGF-1 and IGF-1R protein expression. The suppression of the PI3K/Akt pathway was also demonstrated by examining the ratios of phosphorylated PI3K and Akt, which were reduced in the miR-16 mimic-transfected group compared with the PBS control group. Notably, the IGF-1 expression level and the phosphorylated PI3K ratio were slightly increased in the mimic-NC transfected group (Figure 5C).
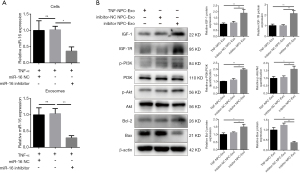
Transfection of NPCs with miR-16 inhibitor decreases exosomal miR-16 levels and blocks the pro-apoptotic effects via the activation of the PI3K/Akt pathway
To further validate the role played by exosomal miR-16 in the induction of NPC apoptosis, miR-16 inhibitors were transfected into NPCs following pretreatment of 10 ng/mL TNF-α for 36 h. The levels of miR-16 in both cells and exosomes were detected by qRT-PCR. miR-16 inhibitors successfully diminished the increased miR-16 level induced by TNF-α treatment in both NPCs and exosomes (Figure 6A). Western blot analysis indicated that NPCs incubated with these exosomes expressed higher protein levels of both IGF-1 and IGF-1R. The downstream PI3K/Akt signaling activity was partially recovered. In addition, the decreased level of the apoptotic marker Bax and the increased level of the anti-apoptotic marker Bcl-2 were also observed (Figure 6B).
Herniated human NP tissues expressed a high level of miR-16, low levels of IGF-1 and IGF-1R, and was characterized by high apoptotic activity
Because it is not possible for validating our in vitro findings in live human discs, we collected 7 NP samples from extrusion and sequestration patients and compared the expression levels of miR-16, IFG-1, IFG-1R, and apoptotic markers between herniated and non-herniated NP tissue. As shown in Figure 7A, the herniated NP tissue from an extrusion patient presented a relatively clear boundary from non-herniated tissues in both the MRI and surgical specimens. The RNA extraction and qRT-PCR detection of the four previously examined miRNAs, miR-452-5p, miR-190a-5p, miR-301a-3p, and miR-16-5p, were performed separately in both herniated and non-herniated tissues. Among these miRNAs, only the expression of miR-16-5p was significantly increased in the herniated NP tissue compared with the non-herniated tissue (Figure 7B).
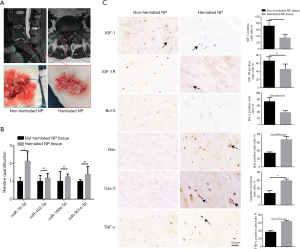
As shown in Table S1, the samples we collected in this study were all classified as Pfirrmann Grade IV or V, which could be considered advanced stages of disc degeneration (20). We performed immunohistochemistry to assess the expression levels of IGF-1, IGF-1R, and TNF-α, as well as protein markers of apoptotic activity in both herniated and non-herniated NP tissues. Both IGF-1 and IGF-1R were expressed at lower levels in herniated NP tissues compared with non-herniated NP tissues. In contrast, TNF-α had elevated expression in herniated NP tissues compared with non-herniated NP tissues. In addition, significantly increased expression levels of the apoptotic markers, Bax and caspase-3, as well as reduced expression levels of the anti-apoptotic marker Bcl-2 were observed in herniated NP samples compared with those in non-herniated NP samples (Figure 7C).
Discussion
Disc degeneration disease has long been considered to be associated with an inflammatory process. The expression levels of proinflammatory cytokines, including TNF-α, IL-1β, and lipopolysaccharide (LPS), are markedly increased in both human and animal models of degenerated disc disease compared with those in normal controls (21,22). Proinflammatory cytokines, particularly TNF-α, can induce apoptosis of NPCs within discs, increasing the expression of proteases responsible for matrix degradation, which can accelerate the structural and biological deterioration of the intervertebral disc (23).
Exosomes are nano-sized particles that function as important mediators of intercellular communications. Interestingly, the activation of exosome release has been reported in cells under inflammatory conditions. The exosome quantity derived from TNF-α-stimulated human NPCs increased significantly compared with those derived from normal NPCs (13). In the present study, we found that the release of exosomes from rat NPCs was increased by TNF-α pretreatment. Such exosomes could induce apoptosis of normal NP cells via transmitting miR-16, which interacted with the 3'-UTR of IGF-1 and IGF-1R mRNA.
Our previous study reported that the activation of autophagy promoted normal exosome release from NPCs via intercellular transportation associated with the Rho-associated coiled-coil containing protein kinase 2 (ROCK2)/RhoC signaling pathway (24). However, the mechanism underlying the inflammation induced-exosome release from NPCs remained unclear. The activation of small GTPases, including the Rho family members Rab and ADP-ribosylation factor 6 (ARF6), are associated with exosome release (25). Interestingly, ARF6 has been reported to be activated by proinflammatory cytokines, such as IL-1β, and has also been reported to promote miRNA transportation into exosomes via the downstream Exportin-5 axis (26). Therefore, the underlying mechanism through which inflammation promotes exosome release from NPCs and whether small GTPases or other potential factors, such as RILP, participate in this process requires further investigation.
Exosome release serves as a method through which cells discard excess cellular components; therefore, we examined whether inflammation-induced exosomes could have negative effects on recipient cells. In our present study, incubation with TNF-Exos induced the apoptosis of normal NPCs. This phenomenon was verified by the western blot detection of increased apoptosis marker expression, including cleaved caspase-3 and Bax, and the decreased expression of the anti-apoptotic marker Bcl-2. In addition, the flow cytometry result also indicated an increased apoptosis rate in NPCs co-cultured with inflammation-induced exosomes.
IGF-1 has been reported to be involved in potent apoptosis inhibition pathways in multiple cell types, including NPCs. The PI3K/Akt signaling pathway downstream of IGF-1 also has anti-apoptotic effects and regulates cell proliferation (27,28). Therefore, we attempted to determine whether IGF-1 was involved in exosome-induced NPC apoptosis. After incubation with TNF-Exos, IGF-1 protein levels in NPCs significantly decreased.
miRNAs are important exosomal cargos that function as negative gene expression regulators capable of regulating multiple targets. We searched for potential exosomal miRNAs that might regulate IGF-1 using two well-established websites that perform miRNA target prediction: TargetScan and miRDB. The cross-section between the two predicted-target lists was used for further study. We picked the top 4 candidate targets based on the prediction scores provided by both websites and analyzed their levels using qRT-PCR in exosomes and NPCs. The qRT-PCR data indicated that only miR-301a and miR-16 were significantly abundant in both TNF-α-stimulated NPCs and their exosomes.
In many cases, exosomes contain much higher levels of miRNAs than the cells these exosomes derive from. In our present study, TNF-α-pretreatment significantly increased the expression levels of miR-16 and miR-301a in both cells and exosomes. However, intercellular miR-16 was expressed approximately 2-fold higher in TNF-α-stimulated cells than in control cells, whereas exosomal miR-16 increased by nearly 4-fold in the stimulated group compared with that in the control group. In contrast, miR-301a did not show a similar trend, and the expression levels displayed similar fold-changes between the stimulated group and the control group. These findings suggested that exosomes tended to selectively contain increased levels of miR-16 under inflammatory conditions relative to the intercellular expression level. However, further efforts should be made to investigate the underlying mechanisms associated with this selective loading of exosomal miRNA.
Recently, RILP was reported as being responsible for selective exosomal miRNA cargo loading, in addition to its promotional effects on exosome release (20). In addition, a small GTPase, ARF6, which has been reported to act as a regulator of exosome release, was also shown to participate in the exosomal loading of miRNAs via the downstream effector Exportin-5 (26). Whether RILP or the ARF6/Exportin-5 axis also plays similar roles in NPCs under inflammatory conditions should be explored.
We opted to focus on miR-16 and attempted to gain a general understanding of its biological effects, based on the findings of previous studies and the miRNA target-prediction tools. Interestingly, when we examined the cross-sectional list of predicted miR-16 targets according to TargetScan and miRDB, we discovered that IGF-1R contained a conserved miR-16 binding site within the 3’UTR. Interestingly, previous studies had reported the regulatory relationship between miR-16 and IGF-1R. It was reported that miR-16 had an inhibitory effect on the proliferation and metastasis of hepatocellular carcinoma and osteosarcoma cell lines (27,28).
Using a dual-luciferase reporter assay, we validated the functionality of the miR-16 binding sites in the 3’UTRs of both IGF-1 and IGF-1R mRNA.
Due to the regulatory effects of miR-16 on both IGF-1 and IGF-1R, we examined whether this coincidence would result in a unique and potent effect on the induction of NPC apoptosis.
We constructed miR-16 mimic-transfected NPCs and isolated the exosomes released from these cells. The qRT-PCR detection of both exosomal and intracellular miR-16 expression levels indicated the successful transfection of the miR-16 mimics. Exosomes derived from miR-16 mimic-transfected NPC significantly repressed the expression of IGF-1 and IGF-1R and inhibited the downstream PI3K/Akt pathway in normal NPCs. To further validate the function of exosomal miR-16 derived from TNF-α-pretreated NPCs, we transfected either miR-16 inhibitors or inhibitor-NC in TNF-α-pretreated NPCs and collected the exosomes from these cells. The exosomal miR-16 induced-repression of IGF-1/IGF-1R was successfully rescued by transfection with miR-16 inhibitors. The activity of the downstream PI3K/Akt pathway was also rescued, which repressed exosome-induced NPC apoptosis.
However, these in vitro findings could not be validated in vivo. Limitations include a lack of techniques capable of facilitating the direct observation of exosomes or exosomal miRNA transportation in vivo and the difficulty identifying precisely which groups of NPCs are under inflammatory conditions. Single-cell sequencing is a newly developed technique that has recently allowed researchers to investigate the heterogeneity of cells within tissues and accurately analyze changes in cellular physiological activities and gene expression at the single-cell level.
Recently, a single-cell sequencing study of rat intervertebral discs identified three groups of cells inside rat discs, including NPCs, AF cells, and stem-like cells, revealing the heterogeneous cell clusters in rat discs (29). This technique could be used in the future to identify different groups of NPCs under inflammatory conditions to analyze their differential responses to external stimuli. The heterogeneity of NPCs in a proinflammatory microenvironment may be fundamental to the cell-to-cell communications that occur between different NPCs.
We also attempted to associate our in vitro findings with clinical observations. Spinal MRI has been considered to represent one of the most important indicators of IDH. In many cases, the pathological disc can be viewed as two parts: herniated and non-herniated. On MRI imaging, non-herniated NP tissue often present with enhanced T2-weighted signal intensity compared with herniated tissue. Previous studies reporting on observations between herniated and non-herniated NP tissues indicated differences in the matrix organization, NPC apoptosis ratio, and other pathological characteristics between these two segments (30).
We wondered whether herniated NP tissues might have negative effects on non-herniated tissues. However, determining whether exosome-mediated cell-to-cell communications participate in this process in vivo is impossible using currently available techniques. The immunohistochemical analysis of both herniated and non-herniated NP tissues indicated the reduced activity of the IGF-1/IGF-1R signaling pathway and increased apoptosis activity in herniated tissues comparing with non-herniated tissues. In addition, the expression levels of miR-16 and TNF-α were also increased in the herniated tissues. These in vivo findings indirectly supported our in vitro results in rats.
In addition to questions that require further study, our work has some limitations. First, only rat NPCs were tested in this study. We searched and validated the miRNA-mRNA regulation relationship only in rat NPCs but not in human NPCs. Therefore, the conclusion we drew in the present study might be somehow different in human NPCs. Moreover, the limited availability of normal human NPCs restricted the clinical relevance of our work. Moreover, NPCs that have been cultured from different donors have been very heterogeneous. The inflammatory microenvironments associated with donor discs were unpredictable and uncontrollable.
The SD rats we used in this study were all raised under the same conditions, were of similar ages, and were of the same gender, which ensured access to a stable source of NPCs to perform this study.
Our findings comparing herniated and non-herniated human NP tissues showed different levels of IGF-1/IGF-1R pathway activity and varying TNF-α levels, which indicated that the inflammatory microenvironment might differ between the two segments of the same degenerated disc, which may be a fundamental reason for the necessity of exosome-based cell-to-cell communications within intervertebral discs.
However, we didn’t perform in vivo animal experiment in the present study. The effect of exosomes derived from abnormal NPCs in rat intervertebral discs awaited further study by intra-disc injection of such exosomes and analyzation of the following imaging examinations as well as molecular biological experiments.
Further study may also take advantage of single-cell sequencing to explore the heterogeneity of NPC under inflammatory conditions.
Conclusions
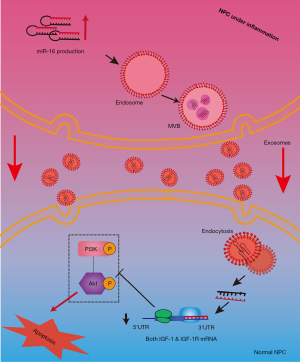
In conclusion, the proinflammatory factor TNF-α was able to stimulate the release of exosomes including miR-16 from NPCs in rats, which further induced the apoptosis of normal NPCs through repressing the anti-apoptotic IGF-1/IGF-1R pathway (Figure 8). Our work revealed a new mechanism through which inflammation and exosomes participate in NPCs apoptosis and suggested that cell-to-cell communications mediated by exosomes played important roles in the maintenance of disc homeostasis.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 81771501 and No. 81701370), Natural Science Foundation of Shanghai (21ZR1412300), “Technology Innovation Action Plan” of Shanghai Science and Technology Commission (21S119 02700), Shanghai “Rising Stars of Medical Talent” Youth Development Program (Youth Medical Talents – Specialist Program, [2020]087) and Shanghai Talent Development Fund (2020067).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-227
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-227
Peer Review File: Available at https://dx.doi.org/10.21037/atm-21-227
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-227). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Research Ethics Committee of Zhongshan Hospital, Fudan University (No. Y2021-228). All tissues were used according to ethical guidelines of the Declaration of Helsinki (as revised in 2013), and the written informed consent for scientific research was obtained from all patients. All animal experiments were carried out in accordance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals. The animal experiments performed in this study were approved by the Animal Care and Use Committee of Fudan University (Shanghai, China, No. Y2021-228).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- . GBD 2017 Disease and Injury Incidence and Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018;392:1789-858. [Crossref] [PubMed]
- Roughley PJ. Biology of intervertebral disc aging and degeneration: involvement of the extracellular matrix. Spine (Phila Pa 1976) 2004;29:2691-9. [Crossref] [PubMed]
- Roberts S. Disc morphology in health and disease. Biochem Soc Trans 2002;30:864-9. [Crossref] [PubMed]
- Bertram H, Nerlich A, Omlor G, et al. Expression of TRAIL and the death receptors DR4 and DR5 correlates with progression of degeneration in human intervertebral disks. Mod Pathol 2009;22:895-905. [Crossref] [PubMed]
- Johnson ZI, Schoepflin ZR, Choi H, et al. Disc in flames: Roles of TNF-α and IL-1β in intervertebral disc degeneration. Eur Cell Mater 2015;30:104-16; discussion 116-7. [Crossref] [PubMed]
- Bachmeier BE, Nerlich AG, Weiler C, et al. Analysis of tissue distribution of TNF-alpha, TNF-alpha-receptors, and the activating TNF-alpha-converting enzyme suggests activation of the TNF-alpha system in the aging intervertebral disc. Ann N Y Acad Sci 2007;1096:44-54. [Crossref] [PubMed]
- Brener A, Sagi L, Shtamler A, et al. Insulin-like growth factor-1 status is associated with insulin resistance in young patients with spinal muscular atrophy. Neuromuscul Disord 2020;30:888-96. [Crossref] [PubMed]
- Osada R, Ohshima H, Ishihara H, et al. Autocrine/paracrine mechanism of insulin-like growth factor-1 secretion, and the effect of insulin-like growth factor-1 on proteoglycan synthesis in bovine intervertebral discs. J Orthop Res 1996;14:690-9. [Crossref] [PubMed]
- Okuda S, Myoui A, Ariga K, et al. Mechanisms of age-related decline in insulin-like growth factor-I dependent proteoglycan synthesis in rat intervertebral disc cells. Spine (Phila Pa 1976) 2001;26:2421-6. [Crossref] [PubMed]
- Pratsinis H, Kletsas D. PDGF, bFGF and IGF-I stimulate the proliferation of intervertebral disc cells in vitro via the activation of the ERK and Akt signaling pathways. Eur Spine J 2007;16:1858-66. [Crossref] [PubMed]
- Pratsinis H, Constantinou V, Pavlakis K, et al. Exogenous and autocrine growth factors stimulate human intervertebral disc cell proliferation via the ERK and Akt pathways. J Orthop Res 2012;30:958-64. [Crossref] [PubMed]
- Liao Z, Luo R, Li G, et al. Exosomes from mesenchymal stem cells modulate endoplasmic reticulum stress to protect against nucleus pulposus cell death and ameliorate intervertebral disc degeneration in vivo. Theranostics 2019;9:4084-100. [Crossref] [PubMed]
- Cheng X, Zhang G, Zhang L, et al. Mesenchymal stem cells deliver exogenous miR-21 via exosomes to inhibit nucleus pulposus cell apoptosis and reduce intervertebral disc degeneration. J Cell Mol Med 2018;22:261-76. [Crossref] [PubMed]
- Zhu G, Yang X, Peng C, et al. Exosomal miR-532-5p from bone marrow mesenchymal stem cells reduce intervertebral disc degeneration by targeting RASSF5. Exp Cell Res 2020;393:112109 [Crossref] [PubMed]
- Zhang J, Zhang J, Zhang Y, et al. Mesenchymal stem cells-derived exosomes ameliorate intervertebral disc degeneration through inhibiting pyroptosis. J Cell Mol Med 2020;24:11742-54. [Crossref] [PubMed]
- Zhu L, Shi Y, Liu L, et al. Mesenchymal stem cells-derived exosomes ameliorate nucleus pulposus cells apoptosis via delivering miR-142-3p: therapeutic potential for intervertebral disc degenerative diseases. Cell Cycle 2020;19:1727-39. [Crossref] [PubMed]
- Zhang QC, Hu SQ, Hu AN, et al. Autophagy-activated nucleus pulposus cells deliver exosomal miR-27a to prevent extracellular matrix degradation by targeting MMP-13. J Orthop Res 2021;39:1921-32. [Crossref] [PubMed]
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4:e05005 [Crossref] [PubMed]
- Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res 2020;48:D127-31. [Crossref] [PubMed]
- Pfirrmann CW, Metzdorf A, Zanetti M, et al. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine (Phila Pa 1976) 2001;26:1873-8. [Crossref] [PubMed]
- Du J, Pfannkuche JJ, Lang G, et al. Proinflammatory intervertebral disc cell and organ culture models induced by tumor necrosis factor alpha. JOR Spine 2020;3:e1104 [Crossref] [PubMed]
- Mern DS, Walsen T, Beierfuß A, et al. Animal models of regenerative medicine for biological treatment approaches of degenerative disc diseases. Exp Biol Med (Maywood) 2021;246:483-512. [Crossref] [PubMed]
- Song XX, Jin LY, Li XF, et al. Substance P Mediates Estrogen Modulation Proinflammatory Cytokines Release in Intervertebral Disc. Inflammation 2021;44:506-17. [Crossref] [PubMed]
- Hu SQ, Zhang QC, Meng QB, et al. Autophagy regulates exosome secretion in rat nucleus pulposus cells via the RhoC/ROCK2 pathway. Exp Cell Res 2020;395:112239 [Crossref] [PubMed]
- van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 2018;19:213-28. [Crossref] [PubMed]
- Clancy JW, Zhang Y, Sheehan C, et al. An ARF6-Exportin-5 axis delivers pre-miRNA cargo to tumour microvesicles. Nat Cell Biol 2019;21:856-66. [Crossref] [PubMed]
- Cheng B, Ding F, Huang CY, et al. Role of miR-16-5p in the proliferation and metastasis of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci 2019;23:137-45. [PubMed]
- Chen L, Wang Q, Wang GD, et al. miR-16 inhibits cell proliferation by targeting IGF1R and the Raf1-MEK1/2-ERK1/2 pathway in osteosarcoma. FEBS Lett 2013;587:1366-72. [Crossref] [PubMed]
- Wang J, Huang Y, Huang L, et al. Novel biomarkers of intervertebral disc cells and evidence of stem cells in the intervertebral disc. Osteoarthritis Cartilage 2021;29:389-401. [Crossref] [PubMed]
- Lama P, Le Maitre CL, Dolan P, et al. Do intervertebral discs degenerate before they herniate, or after? Bone Joint J 2013;95-B:1127-33. [Crossref] [PubMed]


