
Identification and validation of a metabolism-related model and associated with tumor-infiltrating lymphocytes in p53 mutant lung adenocarcinoma patients
Introduction
Lung adenocarcinoma (LUAD), is the most common histological subtype of lung cancer, and has an average 5-year survival rate of only 15% (1,2). Thus, understanding the biology of lung cancer, which is a molecularly heterogeneous disease, is crucial to the development of effective therapies (3). Indeed, lung cancer treatment has developed from the empirical use of physician-recommended cytotoxic therapy towards personalized medicine. In this approach, subsets of patients are treated according to the genetic changes in their tumors and the status of programmed death ligand-1 (PD-L1), which predict the responsiveness to targeted therapies and immune checkpoint blockade, respectively (4). Chemotherapy still represents an effective, evidence-based treatment option for lung cancer (5). In recent studies, the tumor mutational burden (TMB) has been proposed as a distinct biomarker of checkpoint blockade therapy responses (6). This suggests the possibility of variables that simultaneously affect two or more predictive factors; identifying models based on such variables would improve predictive value and ultimately therapeutic outcomes (6).
The identification of somatic mutations that activate specific oncogenic drivers in LUAD has transformed the treatment of non-small cell lung carcinoma. However, it remains unclear how the p53 mutant, which occurs frequently in lung cancer, mediates specific immune-related pathways to produce substantial effects on the tumor microenvironment (TME) (7). In lung cancer, p53 mutation leads to the transcription of downstream target genes involved in the formation of intratumor heterogeneity and may contribute to tumor progression, potentially bearing relevant predictive implications for treatment in many LUAD cases (8). Given that activation of specific oncogenic pathways has broad effects on gene expression, it is reasonable to suggest that the genetic make-up of cancer cells likely has major effects on the TME by driving specific immunoescape.
In the dynamic immune system, metabolic reprogramming induces substantial parallel changes that mediate immunological functions and responses. Recently, glucose metabolism has been associated with the phenotype and activation of immune cells, including macrophage polarization (9), dendritic cells (10), and CD8+ T cells (11). Since immune cells are crucial to TME signals, the various metabolic configurations during tumor progression are of great interest, particularly in patients whose tumors harbor activating p53 mutations.
In the present study, in order to assess the potential associations between unique immune cell compositions and metabolic reprogramming, and thereby identify potential biomarkers, we used an integrative approach that incorporated analysis of the TMB, tumor-associated immune cells, and a metabolic model of the TME in LUAD harboring the p53 mutation. We also validated the prediction model in other independent datasets and wetlab experiments. We present the following article in accordance with the TRIPOD reporting checklist (available at https://dx.doi.org/10.21037/atm-21-3234).
Methods
Study design
This was an observational study that made use of transcriptomic and somatic mutation data. We developed a model for estimating the tumor metabolic risk score (TMRS) of all LUAD samples (n=495) from The Cancer Genome Atlas (TCGA; https://tcga-data.nci.nih.gov, downloaded on 10/09/2020), and evaluated the comprehensive immune landscape by estimating the immune cell composition of the TME. The samples were divided into two distinct clusters based on p53 mutation, and the characteristics of each sample in each cluster, including differences in the TMRS and enrichment of immune cell types, were investigated. The prognostic value of the TMRS and metabolism-related genes was evaluated, and correlations between the TMRS and immune cells were derived. Finally, the results were validated in an independent Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo) dataset (GSE72094, n=442), as well as an immunohistochemistry (IHC) assay, real time-polymerase chain reaction (RT-PCR), and western blot.
Pan-cancer computational analyses
Collection and procession of TCGA datasets
For the 495 TCGA LUAD samples, somatic mutations were determined from Mutation Annotation Format (MAF) files using the VarScan2 algorithm with the default parameters; preprocessed RNA sequencing (RNA-seq) data (level 3) was generated using the Illumina HiSeq 2000 RNA Sequencing Version 2 platform. The reference gene transcript set was the Genome Reference Consortium Human Build 38 patch release 12 (GRCh38.p12) annotation file downloaded from the Ensembl genome browser (http://asia.ensembl.org/biomart), which was used to convert gene symbols. To prepare the expression data profiles for further analysis, count-type RNA-seq data were normalized using the voom method (12) (variance modeling at the observational level), with count data converted to values more similar to those resulting from microarrays.
Construction of p53-related metabolism signatures
The TMRS was compared with gene signatures representing metabolism. Seventy metabolism-related gene sets were obtained from the relevant Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways database (13), which included 186 genes in total, via the Gene Set Enrichment Analysis (GSEA) website (http://software.broadinstitute.org/gsea/downloads).
To assess whether distinct patterns of tumor metabolism were caused by p53 mutations in LUAD and whether these patterns were associated with patient survival, metabolism signatures were obtained as follows. Differentially expressed metabolic genes associated with the p53 mutant were selected using the edgeR package (https://bioconductor.org/packages/edgeR). Significantly differentially expressed genes were defined as where a false discovery rate (FDR) cutoff value of 0.05 was met and |log2 fold change value| was ≥1.
Important variables possibly correlated with the p53 mutant could potentially be excluded when confounding factors were controlled for when variables from differential analyses were selected. To address this issue, LASSO logistic regression models were fitted via the estimated likelihood of deviance with the R package “glmnet” (https://glmnet.stanford.edu). The LASSO model was established depending on the coefficients of the features. We employed K-fold cross-validation with K=10 and the best tuning parameter value with the minimum value of lambda. Based on the coefficients in the LASSO regression model formula, the TMRS of every patient was calculated to obtain the metabolic risk score. To provide comparability of LUAD patients between the high and low TMRS score subsets, the relative metabolic risk scores were scaled according to the median value.
Function annotation and gene set enrichment analysis
The differentially expressed genes between high and low TMRSs were clustered into various KEGG pathway ontologies using the ClueGO Cytoscape plug-in for the visualization of non-redundant biological terms for large clusters of genes in a functionally grouped network. GSEA was performed to identify potential biological processes (BPs) by categorically labelling the samples according to the high and low TMRS type. We tested approximately 1,500 gene sets from the molecular signatures databases (c5.bp.v7.0.symbols) using GSEA v4.1 based on JAVA v8.0 script. The number of random sample permutations was set at 1,000 and the selection threshold was based on the value of the FDR q value <0.05 as a cutoff for inclusion.
Nomogram construction and validation
Clinicopathology (age and gender), p53 mutation status, and TMRS were selected to develop a nomogram via the multivariable Cox proportional hazard model in rms R package (https://hbiostat.org/R/rms). In the multivariate analysis, the nomogram of combined potential risk variables was applied to visualize the predicting survival efficiency. The 1- and 3-year discriminations of the nomogram were validated by Harrell’s concordance index (C-index) and calibration curves. Decision curve analysis (DCA) was performed to show clinical usefulness by calculating the net benefit of the nomogram (14).
TME landscape mapping and clustering
To visualize immune-related cell heterogeneity in the TME, two machine learning methods of gene expression profiles were used: the Estimation of STromal and Immune cells in Malignant Tumors using Expression data (ESTIMATE) (15) and CIBERSORT (16) algorithms (https://cibersort.stanford.edu). First, to obtain the tumor purity of each patient, we used the ESTIMATE algorithm, which reveals the infiltration of tumor and stromal cells. Samples were considered eligible for further analysis when the percentage composition of tumor purity was >0.6. Second, to assess the proportions of 22 tumor-infiltrating lymphocytes in LUAD tissues, “1,000 permutations” was applied as a parameter in CIBERSORT, with the leucocyte gene signature matrix (termed LM22) used as a reference (17). CIBERSORT derives an empirical P value (with a cutoff of <0.05 chosen), which represents the reliability of the deconvolution results. In each sample, the sum of all 22 estimated immune cell type fractions was equal to 1.
The LM22 matrix with 547 genes can be used to distinguish the following 22 human immune cell phenotypes: naive and memory B cells; naive CD4+ T and CD8+ cells; regulatory T cells (Tregs); resting and activated CD4+ memory T cells; follicular helper T cells; γδ T cells; resting and activated mast cells; resting and activated natural killer cells; resting and activated dendritic cells; M0, M1, and M2 macrophages; plasma cells; monocytes; eosinophils; and neutrophils.
Validation in an independent cohort
To verify the associations found among the immune landscape, tumor metabolism, and prognosis, additional analyses were performed in an independent LUAD cohort (GSE72094). The normalized gene expression data of 442 LUAD samples were downloaded from GSE72094. For comparison, the TMRS and immune-related cell proportions of GSE72094 were calculated by the same methods used for TCGA data. The association between GSE72094 variables, including TMRS, immune cells, and overall survival (OS), were analyzed to validate TCGA dataset.
Validation in LUAD cell lines
Cell culture and transient transfections
A549 (p53Wild type, p53Wt) and H1299 (p53Null) LUAD cell lines were routinely cultured in RPMI 1640 medium (GIBCO Thermo Fisher Scientific, MA, USA) containing 10% fetal bovine serum (FBS, Israel), 100 units/mL penicillin, and 100 mg/mL streptomycin (Solarbio, Beijing, China) in an incubator at 37 °C with 5% CO2. The cDNAs for wild type p53 and dominant negative mutant p53 (R248W) were purchased from GenePharma Co, Ltd (Shanghai, China) and transiently transfected using jetPRIME® Transfection Reagent (Polyplus Transfection, Illkirch, France) according to the manufacturer’s protocol, as described previously (18).
Western blotting
Equal amounts of total protein from cells (30 µg) were separated by SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore Immobilon-P, MA, USA). The membranes were incubated at 4 °C overnight with primary antibodies against mannosyl (α-1,3-)-glycoprotein β-1,2-N-acetylglucosaminyltransferase 5B (MGAT5B, ab165538, Abcam, UK), aldehyde dehydrogenase 3A1 (ALDH3A1, ab76976, Abcam, UK),and p53 (#2527, Cell Signaling Technology, USA). After washing, goat anti-mouse/rabbit immunoglobulin G (IgG) antibody was added for 1 h at room temperature. Antibody-antigen binding was detected by high-sig ECL western blotting substrate and visualized by the Tanon 5500 imaging system (Shanghai, China). The protein loading variation was normalized by a-tubulin or GAPDH (ab8245, Abcam, UK). Blot density was analyzed by NIH ImageJ software (Bethesda, MD, USA).
RT-PCR
Total RNA samples were extracted from cells using Trizol reagent (Life Technologies, Carlsbad, CA, USA) using a TissueLyser II (Retsch, Newtown, PA, USA). Extracted RNA samples were subjected to clean-up using 75% ethanol and then reverse transcribed by Prime Script RT reagent Kit (Takara, Dalian, China), as described previously (18). Real-time fluorescence detection was performed using SYBR Premix Ex Taq mix (Takara, Dalian, China) with 5 µg cDNA and 0.1 µM primers on QuantStudio 6 Flex Real-time PCR System (Applied Biosystems, Life Technologies). The thermal cycling parameters were as follows: 95 °C for 10 min, 40 cycles of 95 °C for 15 s, and 60 °C for 1 min, followed by melting curve analysis. The qPCR product was resolved by agarose gel electrophoresis to ensure the correct amplicon length. The primers were designed using Primer Express 4 (Applied Biosystems) and synthesized by Takara Co., Ltd. (Takara, Dalian, China). The following sets of primers were used: MGAT5B (forward 5'-ATCCGCACAGAAGTGATGGG-3' and reverse 5'-CAGCGATGTCGGAGACGTT-3'), ALDH3A1 (forward 5'-TGGAACGCCTACTATGAGGAG-3' and reverse 5'-GGGCTTGAGGACCACTGAG-3'), p53 (forward 5'-CAGCACATGACGGAGGTTGT-3' and reverse 5'-TCATCCAAATACTCCACACGC-3'), and β-ACTIN (Forward 5'-CTCCATCCTGGCCTCGCTGT-3' and reverse 5'-GCTGTCACCTTCACCGTTCC-3'). The mRNA levels of β-ACTIN were used for normalization and calculations, as described previously (19).
Validation in LUAD tissues by IHC
Clinical samples for immunohistochemical staining
To validate the target genes’ protein levels, we obtained a total of 10 pairs of primary tumor tissues and corresponding adjacent normal tissues from LUAD patients who underwent surgical resection at the Department of Thoracic Surgery, First Affiliated Hospital of China Medical University, between May 2020 and October 2020. The location of tumor occurrence in 10 patients included six cases of left lung and four cases of the right lung. There were four cases in males and six cases in females. All patients had postoperative pathology-confirmed LUAD, and underwent surgery prior to chemotherapy or radiotherapy. All samples were used for IHC staining. Our study was conducted according to the Declaration of Helsinki (as revised in 2013) and approved by the First Affiliated Hospital of China Medical University’s Ethics Committee. All patients signed an informed consent form.
Histopathology and immunostaining
Tissues were fixed overnight with 4% paraformaldehyde in PBS. After dehydration in graded concentrations of ethanol, the samples were washed with xylene until opaque. They were then embedded in paraffin and sectioned. Sections (5 µm) were used for IHC as described previously (18). Histopathological images were captured using an immunofluorescent microscope (80I, Nikon Corporation, Tokyo, Japan).
Statistical analyses
Data on age, gender, survival time, and status were collected. The clinical endpoint, OS, was defined as the length of time between the date of diagnosis and the date of death by any cause. Survival analysis was performed using an unadjusted univariate Cox proportional hazard model to test the associations between OS and the other variables in the entire cohort. All statistical tests were performed by survminer package (https://rpkgs.datanovia.com/survminer/index.html) and were two-sided, and P values <0.05 were considered statistically significant. The covariate p53 mutation status, gender, age, and TMRS were included in the multivariable Cox proportional hazard model for adjustment. The log-rank P value was calculated, and the Kaplan-Meier survival curves method was applied to depict the survival rate of the groups. Pearson’s correlation test was used to obtain correlation coefficients from correlations between variables in the different groups.
Results
Distinct metabolism phenotypes in p53 mutant LUAD
The flow diagram in Figure 1A depicts the process of identification and validation of deregulated metabolic genes in LUAD, as well as further correlation analysis of clinicopathological features, p53 mutants, and immune cells.
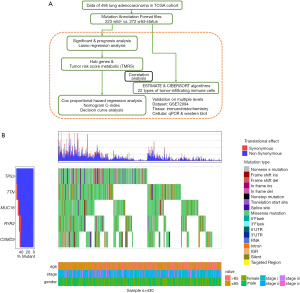
Of the 495 samples, somatic mutations were most frequently located in TP53 [observed in 272 (54.9%) samples with various mutation types (mostly missense mutations)] (Figure 1B). The characteristics of 495 patients with p53 mutation status in the present study from TCGA cohort are displayed in the Table 1, including the patients’ age at diagnosis, gender, and lung cancer clinical stage. There was no significant difference were found between p53 wild and mutant status in the LUAD patients. Given that TP53 was identified as a common oncogenic driver of mutations, we assessed the influence of p53 mutation on metabolic genes.
Full table
Identification of a tumor metabolic gene model for prognostic prediction
To distinguish tumor metabolic patterns between the mutant and wild-type p53 mutation states of LUAD samples, the p53-associated LUAD metabolic genes were selected via comparison analysis, and a LASSO regression model was subsequently constructed to estimate the weighting of metabolic genes and calculate the TMRS for predicting prognosis.
Through distributive difference analysis, 44 differentially expressed metabolism-related genes were identified according to the mutation status of p53 in LUAD (Figure 2A). Of the prognosis-related genes, 31 were involved in the glycose metabolic progress. Through univariate Cox proportional hazard regression analysis, 24 genes related to prognosis were identified (P<0.05). Of these, six genes (ALDH3A1, CD207, CYP2A6, LPL, MGAT5B, NRXN1, STUM, and TPH1) were both differentially expressed and had prognostic value in p53 mutation LUAD. Genes were then selected using the LASSO regression model with logistic regression, and a model was constructed (Figure 2B).
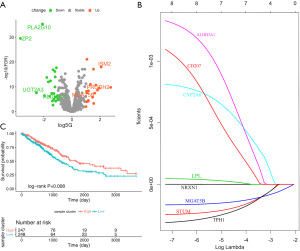
Consequently, a two-gene signature (ALDH3A1: hazard ratio (HR) =0.63; 95% confidence interval (CI): 0.47–0.85; P=0.03; and MGAT5B: HR =1.52; 95% CI: 1.13–2.04; P=0.001) was selected in the model as a classifier, and the TMRS of each patient was formulated with the coefficients of LASSO regression and the corresponding genes’ expression value. The risk score was generated as follows: TMRS = (0.054 × expression level of ALDH3A1) + (−0.0001 × expression level of MGAT5B). Finally, based on the median TMRS, patients were assigned to high- or low-risk groups. The Kaplan-Meier curve showed that patients with a high-TMRS had a significantly shorter OS than low-risk patients (HR =0.39; 95% CI: 0.18–0.85; P=0.008; Figure 2C).
Identification of TMRS-associated biological implications
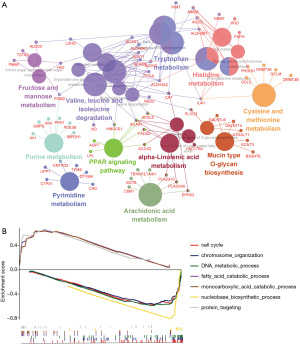
To describe the BPs and mechanisms of hub genes between TMRS subgroups, the gene ontology (GO) functional enrichment analysis was performed using ClueGO and GSEA as a reference. ClueGO was performed to enrich into three significant KEGG pathways (P≤0.05), such as: metabolic pathways, valine, leucine and isoleucine degradation, and tryptophan metabolism (Figure 3A). Furthermore, GSEA analyses indicated that the high-TMRS subgroup was highly enriched in two biological processes: monocarboxylic acid catabolic process and protein targeting. Furthermore, the high-TMRS subgroup was involved in seven pathways: cell cycle, chromosome organization, DNA metabolic process, fatty acid catabolic process, monocarboxylic acid catabolic process, protein targeting, and nucleobase biosynthetic process (Figure 3B).
Construction and evaluation of the nomogram
A nomogram model was used to intuitively visualize the results of the multivariate Cox proportional hazard model, including clinicopathology (age and gender) and genetics (p53 mutation and TMRS). Notably, p53-related TMRS was converted into a categorical variable due to the limitation of continuous predictors. According to the nomogram, p53-related TMRS had the greatest weighting on the prognosis of LUAD, followed by p53 mutation, age, and gender (Figure 4A). In the dynamic nomogram, the total score was ascribed based on each individual factor score that indicated a specific probability of 1- and 3-year survival prognosis (Figure 4B,4C). In addition, the discriminatory power of the nomogram was graphically evaluated using a calibration curve with a C-index of 0.64. The predicted line overlapped well with the reference line, demonstrating that the nomogram performed effectively. In addition, the receiver operating characteristic curves were analyzed, which showed that the predictive accuracy of the nomogram was 0.72 (Figure 4D).

Finally, to determine whether the predictive nomogram was clinically useful, decision curve analysis (DCA) was performed to evaluate the net benefit of the models. As shown in Figure 4E, the decision curve indicated that when the probability of decision-making based on the nomogram is >0.25 and <0.4, it is more beneficial to predict with an all or none-patients scheme.
Correlation between the tumor metabolic risk model and immune response phenotypes
We further illustrated the comprehensive immune landscape in p53 mutant LUAD by assessing the associations with TMRS. Specifically, we tested for associations between the tumor metabolic gene model and tumor-associated immune cells (immune cell composition).
As shown in Figure 5A, CD4 memory resting T cells were more likely to be distributed in higher-level TMRS patients compared with lower-level patients. M1 macrophages were also significantly enriched in higher-level TMRS patients.
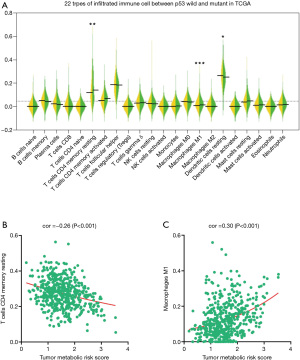
By comparing the TMRS with these two types of immune cell, we represented the correlation between metabolic signatures and tumor immune cells. CD4 memory resting T cells enrichment, based on the percentage of total immune cell content, was negatively correlated with TMRS (r=−0.28, P<0.001; Figure 5B), whereas M1 macrophages were positively correlated with TMRS (r=0.32, P<0.001; Figure 5C). These results suggest that tumor metabolic genes are associated with immune response phenotypes.
Validation in the GSE72094 dataset
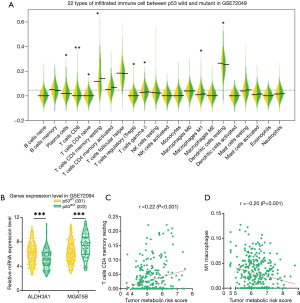
To validate the TCGA LUAD gene set, we also calculated the association between TMRS and the immune landscape, as well as prognostic prediction, using another publicly available independent gene expression dataset (GSE72094), which included 442 LUAD samples. The survival analysis results from GSE72094 were consistent with those of TCGA data (with the same regression coefficients being calculated). As expected, there was a consensus between the results from TCGA and GSE72094 datasets; TMRS was associated with favorable prognosis (HR =0.65; 95% CI: 0.45–0.94; P=0.024). In the TMRS model, ALDH3A1 was significantly associated with favorable OS (HR =0.68; 95% CI: 0.47–0.98; P=0.043), but high levels of MGAT5B were not significantly associated with poor OS.
With filtering based on the ESTIMATE and CIBERSORT algorithms from 331 p53 wild type and 111 p53 mutant samples, 246 p53 wild type and 78 p53 mutant samples were selected for further tumor immune landscape analysis. As shown in Figure 6A, CD4 memory resting T cells and tumor-related macrophages M1 were observed to be differentially expressed between p53 wild and mutant LUAD patients, which was consistent TCGA LUAD cohort. Both ALDH3A1 and MGAT5B were significantly expressed between p53 wild and mutant LUAD patients, which was consistent TCGA LUAD cohort (Figure 6B). There was a negative correlation between CD4 memory resting T cells and the TMRS (r=−0.20, P<0.001; Figure 6C), while M1 macrophages were positively correlated with the TMRS (r=0.22, P<0.001; Figure 6D).
Validation of MGAT5B and ALDH3A1 in p53 mutant LUAD
A dimensional validation was performed to explore the expressions of MGAT5B and ALDH3A1 in the p53 wild and mutant LUAD patients, and the LUAD cell lines.
To investigate whether the expression of MGAT5B and ALDH3A1 is associated with p53 mutations at the cellular level, we transfected wild type p53 cDNA into p53 null cell lines H1299, and delivered p53 mutant type (p53Mt) cDNA into A549 cells, and then detected the status of MGAT5B, ALDH3A1, and p53. As shown in Figure 7A,7B, compared with those in parental or vector-transfected control cells, the protein and mRNA expression levels of MGAT5B increased in H1299 p53Wt cells, while the expression of ALDH3A1 was low. Also, compared with those in control cells, we observed increased ALDH3A1 and decreased MGAT5B in A549 p53Mt cells (Figure 7A,7B).
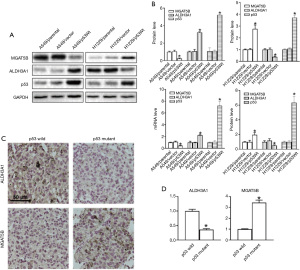
Secondly, IHC analysis of LUAD specimens was preformed to examine the expressions of MGAT5B and ALDH3A1 in p53 wild and mutant LUAD specimens. The results showed that the expression of ALDH3A1 in LUAD patients with p53 mutation was significantly higher than that of patients with p53 wild type (P<0.05). Meanwhile, the expression of MGAT5B was significantly lower than in p53 mutant LUAD tissues (Figure 7C,7D).
Discussion
Tumor cells are hypothesized to reprogram immunity and metabolism in the TME, which may result in immune escape (20). The unique TME may then play a critical role in promoting tumor development, metastasis, and immune evasion (21). We studied the connection between immune cell composition and cancer metabolism to identify the possible roles and mechanisms of the TME in p53 mutation LUAD patients. We developed and validated a gene model of glucose metabolism and tumor-infiltrating immune cells, with transcriptomic data used as an input. MGAT5B and ALDH3A1 genes were involved in the activation of the glycolysis and glycolysis-gluconeogenesis pathways, which promote alternative metabolic rearrangement. We also identified low levels of CD4 memory resting T cells and high M1 macrophage fractions, respectively, in the p53 mutant LUAD TME (based on results from the ESTIMATE and CIBERSORT algorithms). Multidimensional validation subsequently confirmed the reliability of our results. These results indicate that p53 mutation status influences both immunologic microenvironments and glycoses metabolic reprogramming; therefore, these mutations play an essential role in the development of lung cancer.
As a predictor of heterogeneous treatment benefits, we found that the gene signatures significantly outperformed p53 mutation status in the LUAD patient cohort with an independent response. We demonstrated that the TME constitutes a heterogeneous group of tumor-infiltrating lymphocytes, characterized by somatic mutations in the p53 and glycoses metabolic pathways. In recent years, p53 has been found to: (I) play a direct role in innate and adaptive immune responses (22); (II) be involved in regulating the immune checkpoint molecules programmed cell death-1 and its ligand (PD-L1) (23); and (III) to function in the response to antigen-presenting cells, such as dendritic cells, derived from p53 mutations, which may active cytotoxic T-cell recognizing and natural killer cells (24). In oncogene-addicted diseases (harboring EGFR mutations and ALK translocations), the concomitant presence of p53 mutations is associated with resistance to standard tyrosine kinase inhibitors (TKI) treatment. Regarding immunotherapy, a growing body of data is becoming available, suggesting its potential impact (25). Our results showed that an increase in M1 macrophages and a decrease in CD4 memory resting T cells were associated with tumorigenesis of p53 mutant LUAD. Similar results were reported for another subtype of lung cancer, lung squamous cell carcinoma, in which increased infiltration of M1 macrophages was found to be correlated with the p53 mutant (26). Additionally, p53 is a known regulator of macrophage differentiation (27,28). Furthermore, numerous studies have shown that CD4 memory resting T cells play a pivotal role in the immune control of human immunodeficiency virus (HIV) (29) and cancers (30).
We detected metabolic differences between p53 mutation statuses in LUAD within the same transcription dataset. We found that MGAT5B-mediated N glycan biosynthesis and the ALDH3A1-meditated glycolysis-gluconeogenesis metabolic pathway were associated with the p53 mutant and the prognosis of LUAD. MGAT5B is involved in the procession of glycosyl transferase on N-linked glycans, and results in an aberrant glycosylation pattern in cancer cells (31). β(1,6)-branched oligosaccharides, as the products of MGAT5B, are well-known predictors of poor prognosis and decreased survival time in human cancers (32), which is consistent with our findings. In addition, ALDH3A1 has previously been correlated with cigarette smoke in airway epithelial cells (33). The correlation between these two metabolic genes and immune editing in the TME suggests that they should be the focus of further research.
Enhanced glycolysis is a key metabolic route for cancer promotion and metabolism, and is also known as the “Warburg effect” (34). Unlimited proliferation potential and the Warburg effect in cancer cells lead to the production of high levels of lactate in the TME, which further facilitates the progression of tumors and immune escape (35). One important mechanism of immune escape is the glucose competition that occurs between cancer cells and lymphocytes infiltrating tumor tissue. Competition over glucose resources also depicts the role in regulating the proliferation and differentiation of immune cells (36). Tregs may contribute to immune escape mechanisms in the tumor immune response, and are important factors in inducing an antitumor response (37). During T-cell proliferation or differentiation into effector CD4+ T cells, increased glucose metabolism adaptations are required, similar to cancer cells. Coincidently, the glycosylation processing pathways have coevolved with the larger regulatory network that controls T cell activation (35). In the present study, CD4 memory resting T cells and high levels of M1 macrophages were related to N-glycans and ALDN. N-glycans may regulate cell adhesion by contributing to the immune system and tumor metastasis. Consistently, previous studies have reported that the core fucosylation of N-glycan, which is required for T cell receptor signaling with CD4+ T cell activation, is significantly increased in systemic lupus erythematosus patients (38). Upregulated MGAT5 has also been shown to enhance branched N-glycans on T-cell receptors, limiting their capacity by increasing T-cell activation thresholds (39). In one study, MGAT5 was shown to restrict CD8+ T cell activation via IL-10 to promote the establishment of chronic viral infections (40), while another study reported that MGAT5 is involved in regulating macrophage polarization (41). Furthermore, ALDH3A1 has also been found to provoke metabolic reprogramming, leading to immunoevasion in lung cancer (42). Taken together, these results suggest that glucose metabolism could regulate the infiltration of tumor lymphocytic cells and promote tumor progression.
However, the specific roles and mechanisms of these two studied genes in the metabolic and immune reprogramming of the TME remain unclear and this demands further investigation. We also recognize the limitations of our study. Firstly, identifying the entire landscape of infiltrating lymphocytes is difficult because of current limitations in technology and bioinformatics. Further biological experiments and a systematic review could address this limitation. Secondly, although the model was cross-validated by an independent dataset, the roles and mechanisms of hub genes in the metabolic and immune reprogramming of the TME require further exploration in experimental and clinical studies.
Conclusions
Within the scope of our study, we used a multiscale modeling approach to correlate metabolic reprogramming with the tumor-infiltrating lymphocytes of the TME in p53 mutant lung cancer. We identified the MGAT5B and ALDH3A1, also combined them as metabolic model, would have an association with the prognosis of p53 mutant LUAD patients, and the stability and independence have been verified. We also found that the risk score was related to the immune cell infiltration component (CD4 memory resting T cells and M1 macrophages). Overall, this study provides new insights that will inform future investigation into the complex factors that contribute to the TME.
Acknowledgments
Funding: This work was supported by grants from the National Natural Science Foundation of China (No. 81773524).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-3234
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-3234
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-3234). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the First Affiliated Hospital of China Medical University’s Ethics Committee. The clinical analysis was performed according to the principles of the Helsinki Declaration (as revised in 2013). All patients signed an informed consent form.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A, et al. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018;362:eaar3593 [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C, et al. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Wray NR, Wijmenga C, Sullivan PF, et al. Common Disease Is More Complex Than Implied by the Core Gene Omnigenic Model. Cell 2018;173:1573-80. [Crossref] [PubMed]
- Hellmann MD, Nathanson T, Rizvi H, et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell 2018;33:843-852.e4. [Crossref] [PubMed]
- Turrell FK, Kerr EM, Gao M, et al. Lung tumors with distinct p53 mutations respond similarly to p53 targeted therapy but exhibit genotype-specific statin sensitivity. Genes Dev 2017;31:1339-53. [Crossref] [PubMed]
- Mills EL, Kelly B, Logan A, et al. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell 2016;167:457-470.e13. [Crossref] [PubMed]
- Everts B, Amiel E, Huang SC, et al. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKɛ supports the anabolic demands of dendritic cell activation. Nat Immunol 2014;15:323-32. [Crossref] [PubMed]
- Gubser PM, Bantug GR, Razik L, et al. Rapid effector function of memory CD8+ T cells requires an immediate-early glycolytic switch. Nat Immunol 2013;14:1064-72. [Crossref] [PubMed]
- Law CW, Chen Y, Shi W, et al. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014;15:R29. [Crossref] [PubMed]
- Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353-61. [Crossref] [PubMed]
- Fitzgerald M, Saville BR, Lewis RJ, et al. Decision curve analysis. JAMA 2015;313:409-10. [Crossref] [PubMed]
- Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [Crossref] [PubMed]
- Gentles AJ, Newman AM, Liu CL, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 2015;21:938-45. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Kong W, Fu J, Liu N, et al. Nrf2 deficiency promotes the progression from acute tubular damage to chronic renal fibrosis following unilateral ureteral obstruction. Nephrol Dial Transplant 2018;33:771-83. [Crossref] [PubMed]
- Zheng C, Li X, Ren Y, et al. Coexisting EGFR and TP53 Mutations in Lung Adenocarcinoma Patients Are Associated With COMP and ITGB8 Upregulation and Poor Prognosis. Front Mol Biosci 2020;7:30. [Crossref] [PubMed]
- Choi H, Na KJ, et al. Integrative analysis of imaging and transcriptomic data of the immune landscape associated with tumor metabolism in lung adenocarcinoma: Clinical and prognostic implications. Theranostics 2018;8:1956-65. [Crossref] [PubMed]
- Ngwa VM, Edwards DN, Philip M, et al. Microenvironmental Metabolism Regulates Antitumor Immunity. Cancer Res 2019;79:4003-8. [Crossref] [PubMed]
- Biton J, Mansuet-Lupo A, Pécuchet N, et al. TP53, STK11, and EGFR Mutations Predict Tumor Immune Profile and the Response to Anti-PD-1 in Lung Adenocarcinoma. Clin Cancer Res 2018;24:5710-23. [Crossref] [PubMed]
- Pascual M, Mena-Varas M, Robles EF, et al. PD-1/PD-L1 immune checkpoint and p53 loss facilitate tumor progression in activated B-cell diffuse large B-cell lymphomas. Blood 2019;133:2401-12. [Crossref] [PubMed]
- Castle JC, Uduman M, Pabla S, et al. Mutation-Derived Neoantigens for Cancer Immunotherapy. Front Immunol 2019;10:1856. [Crossref] [PubMed]
- Dong ZY, Zhong WZ, Zhang XC, et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin Cancer Res 2017;23:3012-24. [Crossref] [PubMed]
- Xu F, Lin H, He P, et al. A TP53-associated gene signature for prediction of prognosis and therapeutic responses in lung squamous cell carcinoma. Oncoimmunology 2020;9:1731943 [Crossref] [PubMed]
- Matas D, Milyavsky M, Shats I, et al. p53 is a regulator of macrophage differentiation. Cell Death Differ 2004;11:458-67. [Crossref] [PubMed]
- Lim YJ, Lee J, Choi JA, et al. M1 macrophage dependent-p53 regulates the intracellular survival of mycobacteria. Apoptosis 2020;25:42-55. [Crossref] [PubMed]
- Balasubramaniam M, Pandhare J, Dash C, et al. Immune Control of HIV. J Life Sci (Westlake Village) 2019;1:4-37. [Crossref] [PubMed]
- Borst J, Ahrends T, Bąbała N, et al. CD4+ T cell help in cancer immunology and immunotherapy. Nat Rev Immunol 2018;18:635-47. [Crossref] [PubMed]
- Lange T, Ullrich S, Müller I, et al. Human prostate cancer in a clinically relevant xenograft mouse model: identification of β(1,6)-branched oligosaccharides as a marker of tumor progression. Clin Cancer Res 2012;18:1364-73. [Crossref] [PubMed]
- Gao HJ, Chen YJ, Zuo D, et al. Quantitative proteomic analysis for high-throughput screening of differential glycoproteins in hepatocellular carcinoma serum. Cancer Biol Med 2015;12:246-54. [PubMed]
- Jang AJ, Lee JH, Yotsu-Yamashita M, et al. A Novel Compound, "FA-1" Isolated from Prunus mume, Protects Human Bronchial Epithelial Cells and Keratinocytes from Cigarette Smoke Extract-Induced Damage. Sci Rep 2018;8:11504. [Crossref] [PubMed]
- Schwartz L, Supuran CT, Alfarouk KO, et al. The Warburg Effect and the Hallmarks of Cancer. Anticancer Agents Med Chem 2017;17:164-70. [Crossref] [PubMed]
- Siska PJ, Singer K, Evert K, et al. The immunological Warburg effect: Can a metabolic-tumor-stroma score (MeTS) guide cancer immunotherapy? Immunol Rev 2020;295:187-202. [Crossref] [PubMed]
- Gottfried E, Kreutz M, Mackensen A, et al. Tumor metabolism as modulator of immune response and tumor progression. Semin Cancer Biol 2012;22:335-41. [Crossref] [PubMed]
- Peng M, Yin N, Chhangawala S, et al. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 2016;354:481-4. [Crossref] [PubMed]
- Liang W, Mao S, Sun S, et al. Core Fucosylation of the T Cell Receptor Is Required for T Cell Activation. Front Immunol 2018;9:78. [Crossref] [PubMed]
- Demetriou M, Granovsky M, Quaggin S, et al. Negative regulation of T-cell activation and autoimmunity by Mgat5 N-glycosylation. Nature 2001;409:733-9. [Crossref] [PubMed]
- Smith LK, Boukhaled GM, Condotta SA, et al. Interleukin-10 Directly Inhibits CD8+ T Cell Function by Enhancing N-Glycan Branching to Decrease Antigen Sensitivity. Immunity 2018;48:299-312.e5. [Crossref] [PubMed]
- Yang P, Zhang X, Lin Z, et al. Adoptive transfer of polarized M2c macrophages ameliorates acute rejection in rat liver transplantation. Am J Transl Res 2020;12:2614-26. [PubMed]
- Terzuoli E, Bellan C, Aversa S, et al. ALDH3A1 Overexpression in Melanoma and Lung Tumors Drives Cancer Stem Cell Expansion, Impairing Immune Surveillance through Enhanced PD-L1 Output. Cancers (Basel) 2019;11:1963. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


