Analysis of the expression profile of serum exosomal lncRNA in breast cancer patients
Introduction
Breast cancer (BC) is the most widely diagnosed cancer among women and is the most fatal malignant tumor type, with a morbidity and mortality rate of 25% and 15%, respectively (1). According to the latest data, more than 250,000 patients are diagnosed with BC every year in the United States (2). Although 70% of genes can be transcribed into ribonucleic acids (RNAs), only 2% of these transcripts have been translated into proteins in the human genome. Other transcripts are defined as non-coding RNA and can be divided into two categories. The first is short-chain non-coding RNAs (sncRNAs), which include microRNAs (miRNAs), small interfering RNAs (siRNAs), and small nucleolar RNAs (snoRNAs). The second category includes long-chain non-coding RNAs (lncRNAs) (3). sncRNAs are mainly considered a negative regulator of gene expression, while lncRNAs have been identified as a widely heterogeneous population, and studies of their gene expression have attributed them to the development of many human diseases, including cancer (4,5). Exosomes are membrane vesicles with a diameter of 20–200 nm. They belong to the larger family of extracellular vehicles (EVs) produced in cells and released into the extracellular space (6). Many studies have shown that exosomes are the key mediator of intercellular communication between tumor and stromal cells in local and distant microenvironments (7). Exosomes derived from tumors promote angiogenesis and coagulation, regulate the immune system, reshape the surrounding parenchyma, and jointly support tumor progression (8,9). These exosomes participate in many cellular functions and are considered important cellular communication connectors as they contain various proteins and nucleic acids, including miRNAs and lncRNAs (10,11).
In line with increasing research on the topic, a large amount of evidence shows that the signals transmitted by exo-lncRNAs regulate many types of local and distant receptor cells. In addition, exosome-mediated lncRNA transfer pathways are different in level and type under normal physiological and pathological conditions. Exosomes are extracellular vesicles secreted by different types of cells. Exosomes have become an indispensable promoter of information exchange between cells. More importantly, exosomes play a vital role in various diseases including cancer. Exosomes lncRNAs play a central role in carcinogenesis and cancer progression by regulating tumor growth, metastasis, angiogenesis and chemotherapy resistance. In addition, exosomes lncRNAs play a messenger role in intercellular communication, thus remodeling tumor microenvironment. Their functional relevance in cancer biology suggests that exosome lncRNA may be a promising non-invasive biomarker in future cancer treatment.We used Exo-lncRNA chip technology to detect and compare the differential expression profiles of serum exo-lncRNAs between BC patients and healthy subjects for this study. This was done by screening out the differential exo-lncRNAs and analyzing their targets/possible functional mechanisms through bioinformatics. Through this approach, we hoped to further research on the early diagnosis and treatment of BC. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/atm-21-3483).
Methods
General information
Blood samples were collected from two patients who were diagnosed for the first time with BC at Henan Cancer Hospital (from October 2019 to December 2019). One had early-stage BC (T1), while the other had advanced BC (M1). The pathological type of both patient tumors was confirmed as non-special invasive BC (NST) by a pathologist (T1: early-stage NST; M1: advanced BC with bone metastasis). A further blood sample was collected from a healthy female volunteer. No clinical treatment was given to either BC patient before sample collection. The study was undertaken following the World Medical Association (WMA) Declaration of Helsinki (as revised in 2013), the National Health and Family Planning Commission’s Measures for the Ethical Review of Biomedical Research Involving Human Beings, and other relevant laws and regulations. It was approved by the Medical Ethics Committee of Henan Cancer Hospital. All the subjects gave their consent and signed the relevant consent form.
Instruments and main reagents
Instrument: Agilent 2200 TapeStation Software (Agilent Technologies, CA, USA); Reagents: Qubit (Life Technologies, MA, USA); RNA ScreenTape and RNA Reagent (Agilent Technologies, CA, USA); D1000 ScreenTape and D1000 Reagent (Agilent Technologies, CA, USA); RiboTM Exosome Isolation Reagent (RiboBio, Guangzhou, China); Qubit dsDNA HS Assay Kit (Life Technologies, MA, USA); Agencourt Ampure XP beads (Beckman Coulter, CA, USA); NEB NEXT Ultra RNA Library Prepkit for Illumina (New England Biolabs, MA, USA).
Extraction and identification of serum exosomes
Peripheral venous blood was collected from the subjects with an EDTA-K2 anticoagulation tube. After collection, the sample remained still before being centrifuged. The serum was then collected and packed in sterile EP tubes and temporarily stored at −80 °C. Exosomes were identified by particle size detection and marker protein analysis, and the antibodies used included CD9, CD63, TSGl01, and goat anti-rabbit secondary antibody.
Exosomal RNA extraction and exosomal lncRNA high-throughput sequencing
Exosomal RNAs were extracted following the relevant step-by-step instructions. RNAs in serum exosomes were extracted, and samples were tested by Qubit and Agilent 2200 TapeStation. We then constructed the library following these key steps: (I) RNA fragmentation; (II) first-chain complementary DNA (cDNA) synthesis; (III) synthesis and purification of second-chain cDNA; (IV) end repair and detailing; (V) adapter connection, fragment selection, and purification; (VI) polymerase chain reaction (PCR) amplification and purification. Using the Agilent 2200 TapeStation, the samples then passed a library-quality inspection. This involved preparing the on-machine samples according to the method described in the HiSeq User Guide. Finally, the sequencing was carried out by the Guangzhou Ribobio Biological Company, and the expression data of lncRNAs were analyzed by comparing them with the known gene sequences.
Differential screening
The expression difference between exo-lncRNAs was screened and identified using the following steps: (I) the gene type was set as lncRNA; (II) the expression multiple [log2(Fold change), expression difference multiple] of exosome lncRNAs in the BC group was found to be greater than or equal to 1, or less than or equal to 1.5; (III) exocrine lncRNAs with either high or low expressions were identified in both samples provided by the T1 and M1 BC patients.
Bioinformatics and statistical analysis
Cis/trans target genes were identified in the lncRNAs, and their enrichment functions and signal pathways were analyzed through GO and KEGG analysis. This helped us to predict further the cellular function and signal pathways involved in these abnormally expressed lncRNAs, as well as speculate on the biological function of the lncRNAs. In the prediction of lncRNA-miRNA, miRanda, PITA, and RNAhybrid were used to establish the recognition areas of lncRNA and miRNA.
Results
Exo-lncRNA sequencing results
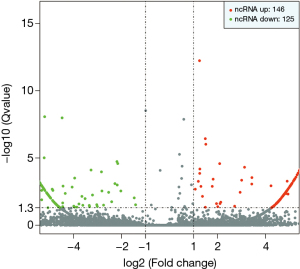
The statistical results of the screening of the differentially expressed exo-lncRNAs are shown in Figure 1. We found that the BC patients had 146 exo-lncRNAs up-regulated and 125 exo-lncRNAs down-regulated when compared with the healthy subject.
The most significantly differentially expressed exo-lncRNAs and a literature review
Through comprehensively comparing the healthy subject with the T1 and M1 BC patients, we obtained 11 significantly different Exo-lncRNAs according to the screening principle. Five of these exosomes displayed a significant increase in the expression levels of lncRNAs, while the other 6 exemplified a significant decrease in expression levels (Table 1). A literature review was performed on these 11 exo-lncRNAs, and 3 of them were identified as VIM-AS1, SNHG8, and ELDR, all of which we further analyzed. VIM-AS1 was noted as up-regulated, while SNHG8 and ELDR were down-regulated.
Table 1
| Lnc-RNA | Gene | Type | Log2 (fold change) | P |
|---|---|---|---|---|
| NR_135133.1 |
|
lncRNA | 4.440055 | 0.00041 |
| NR_102754.1 |
|
lncRNA | 4.369055 | 0.000587 |
| NR_144487.1 |
|
lncRNA | 4.25746 | 0.001 |
| NR_051989.1 |
|
lncRNA | 2.135252 | 0.000289 |
| NR_108060.1 |
|
lncRNA | 1.258204 | 8.34E-07 |
| NR_121667.1 |
|
lncRNA | −1.54641 | 7.07E-24 |
| NR_034010.1 |
|
lncRNA | −3.20761 | 7.01E-05 |
| NR_125380.1 |
|
lncRNA | −3.2679 | 4.17E-07 |
| NR_110426.1 |
|
lncRNA | −4.4345 | 1.11E-05 |
| NR_110278.1 |
|
lncRNA | −5.91017 | 0.000987 |
| NR_135057.1 |
|
lncRNA | −6.71971 | 0.000745 |
Prediction and analysis of lncRNA targets
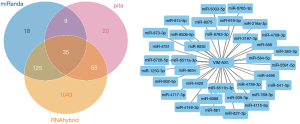
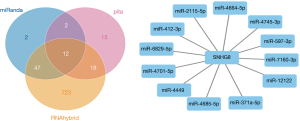
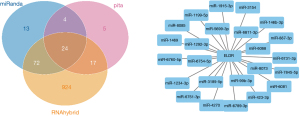
The prediction and detailed information of cis and trans lncRNA targets can be seen in Table 2. The target gene vim of VIM-AS1 was predicted in cis, and the target gene PRSS12 of SNHG8. As for the lncRNA-miRNA prediction analysis target, the results were drawn from three different types of software (miRanda, PITA, and RNAhybrid) and were further screened. The results of which looked at the interaction of the miRNAs in the lncRNAs, with VIM-AS1 predicting 35 target miRNAs, SNHG8 was predicting 12 target miRNAs, and ELDR predicting 24 target miRNAs. Figures 2-4 show the software results.
Table 2
| Transcript | Cis-prediction | Trans-prediction | lncRNA | Chromosome | mRNA | Gene |
|---|---|---|---|---|---|---|
| NR_108060.1 | 1 | 0 | VIM-AS1 | chr10 | NM_003380.3 | VIM |
| NR_034010.1 | 1 | 0 | SNHG8 | chr4 | NM_003619.3 | PRSS12 |
| NR_110426.1 | 0 | 0 | ELDR | – | – | – |
Analysis of GO saliency function enrichment and KEGG pathway saliency enrichment
The functional and signaling pathway enrichment analyses of the GO and KEGG target genes in the lncRNA candidate revealed that VIM, the target gene of VIM-AS1, was enriched with 107 functions in biological processes such as cellular processes, biological regulation, metabolic regulation, macromolecular metabolic processes, stimulation reactions, and others. There were 43 functions enriched in the cell components, which were mainly concentrated in the cells and cytoplasm. In terms of the molecular function, 19 functions were enriched, including adhesion, protein, macromolecule, binding, organic compound, and the complex combination of nucleic acid and protein. PRSS12, a target gene of SNHG8, was biologically enriched with 24 functions, including cellular metabolism, organic metabolism, macromolecular metabolism, gene expression, and protein metabolism. 30 functions were enriched in cell components, mainly concentrated in the cells, cytoplasm, cell membrane, and cell periphery. 12 functions were also enriched in the molecular function; this mainly included catalytic activity, hydrolase activity, receptor activity, molecular sensor, endopeptidase activity, and serine hydrolase activity. KEGG pathway enrichment results also indicated how the VIM protein functions in cancer development through the viral infection signaling pathway and miRNA signaling pathway. The top 10 functions were listed based on the database sample size compared by GO analysis (Tables 3-5).
Table 3
| GO-ID | Term |
|---|---|
| Biological process | |
| GO:0009987 | Cellular process |
| GO:0044699 | Single-organism process |
| GO:0044763 | Single-organism cellular process |
| GO:0065007 | Biological regulation |
| GO:0008152 | Metabolic process |
| GO:0050789 | Regulation of biological process |
| GO:0050794 | Regulation of cellular process |
| GO:0043170 | Macromolecule metabolic process |
| GO:0050896 | Response to stimulus |
| GO:0032501 | Multicellular organismal process |
| Cellular component | |
| GO:0044464 | Cell part |
| GO:0005623 | Cell |
| GO:0005622 | Intracellular |
| GO:0044424 | Intracellular part |
| GO:0043226 | Organelle |
| GO:0043229 | Intracellular organelle |
| GO:0043227 | Membrane-bounded organelle |
| GO:0043231 | Intracellular membrane-bounded organelle |
| GO:0005737 | Cytoplasm |
| GO:0044444 | Cytoplasmic part |
| Molecular function | |
| GO:0005488 | Binding |
| GO:0005515 | Protein binding |
| GO:0097159 | Organic cyclic compound binding |
| GO:1901363 | Heterocyclic compound binding |
| GO:0003676 | Nucleic acid binding |
| GO:0097367 | Carbohydrate derivative binding |
| GO:0003723 | RNA binding |
| GO:0044877 | Macromolecular complex binding |
| GO:0042802 | Identical protein binding |
| GO:0032403 | Protein complex binding |
Table 4
| GOID | Term |
|---|---|
| Biological process | |
| GO:0009987 | Cellular process |
| GO:0044699 | Single-organism process |
| GO:0008152 | Metabolic process |
| GO:0044763 | Single-organism cellular process |
| GO:0071704 | Organic substance metabolic process |
| GO:0044238 | Primary metabolic process |
| GO:0043170 | Macromolecule metabolic process |
| GO:0051179 | Localization |
| GO:0010467 | Gene expression |
| GO:0019538 | Protein metabolic process |
| Cellular component | |
| GO:0044464 | Cell part |
| GO:0005623 | Cell |
| GO:0005622 | Intracellular |
| GO:0044424 | Intracellular part |
| GO:0043226 | Organelle |
| GO:0043227 | Membrane-bounded organelle |
| GO:0005737 | Cytoplasm |
| GO:0016020 | Membrane |
| GO:0044444 | Cytoplasmic part |
| GO:0071944 | Cell periphery |
| Molecular function | |
| GO:0003824 | Catalytic activity |
| GO:0016787 | Hydrolase activity |
| GO:0004872 | Receptor activity |
| GO:0060089 | Molecular transducer activity |
| GO:0070011 | Peptidase activity, acting on L-amino acid peptides |
| GO:0008233 | Peptidase activity |
| GO:0004175 | Endopeptidase activity |
| GO:0008236 | Serine-type peptidase activity |
| GO:0017171 | Serine hydrolase activity |
| GO:0004252 | Serine-type endopeptidase activity |
Table 5
| Signal pathway | ID |
|---|---|
| Epstein-Barr virus infection | hsa05169 |
| MicroRNAs in cancer | hsa05206 |
Discussion
Exosomes are bilayer lipid membrane vesicles with a diameter of about 30–100nm. They contain a variety of protein and nucleic acid components without organelles. They are cup-shaped under electron microscopy and generally have a spherical structure in body fluid. The density range is 1.13–1.19 g/mL (12) in a sucrose density gradient solution. Studies have shown that when compared with healthy subjects, the concentration of exosomes in the plasma of BC patients is significantly increased. This indicates that the number of exosomes in plasma can aid the identification of BC (13). Ewaisha et al. (14) also found that the plasma exosomes of BC patients express specific proteins and RNAs, which play a major role in the occurrence and development of BC. This only further indicates that the contents of exosomes have the potential to be biomarkers for diagnosing BC.
Miao et al. (15) found that the expression level of lncRNA MALATl in the cancer tissue of BC patients was significantly higher than that in the para-cancerous tissue. They also found that the expression level of MALATl in serum samples of BC patients was significantly higher than that in benign breast diseases. In addition, scientists found that lncRNA H19 was significantly expressed in BC serum samples and found that its expression in serum samples after surgery was significantly lower than serum samples taken before surgery. All this research indicates that lncRNAs can be used for the differential diagnosis of BC and has a certain value in the context of prognosis detection. Through a further comparison of the BC patients and the healthy subject, lncRNAs with traditional biomarkers such as CEA and CA-l53 also revealed that lncRNAs had high sensitivity and specificity, even exceeding the sensitivity of traditional ultrasonic diagnostic methods (16). lncRNAs can also be packaged into exosomes and act as messengers in intercellular communication, indicating the further potential for using lncRNAs as a diagnostic and prognostic marker for various other cancers (17,18). The exosome lncRNA Xist-mediated pathway is activated in the early stage of metastatic BC in the brain and may become an effective target for treating brain metastasis (19). The expression of exosome HOTAIR is positively correlated with the state of the receptor tyrosine kinase (RTK) ErbB2 (also known as HER2/neu) in tumor tissues and suggests poor prognosis and chemotherapeutic efficacy (20,21). Exosome H19 can be used as a biomarker for predicting BC and can induce BC resistance (22,23).
VIM-AS1 is a 1.8-kb non-coding RNA. Studies have shown that VIM-AS1 has a hybrid R-loop structure of DNA, shares a bidirectional promoter transcription with VIM mRNA, and can positively regulate Vim expression (24). This is consistent with our predicted target results, but further verification is needed. VIM-AS1 has also been confirmed in human colon cancer cell lines, with its expression being closely related to tumor progression and found to be significantly up-regulated with the progression of tumors. More specifically, the expression of VIM-AS1 in human colon cancer cell lines was up-regulated in lymph node metastasis and vascular invasion tumors. Subsequent in vitro experiments proved that VIM-AS1 played a key role in promoting migration and EMT of colon cancer cells (25), thus further demonstrating the importance of VIM-AS1 in tumorigenesis. The second important exo-lncRNA from our study was SNHG8, which has been noted to play the role of an oncogene in many kinds of tumors, and which was also consistent with our sequencing results. SNHG8 is also involved in tumor drug resistance, angiogenesis, and epithelial-mesenchymal transition, which is consistent with our target prediction and functional analysis. Recent studies have also suggested that a targeted ELDR injection has good therapeutic potential for oral cancer (26). From our findings, we believe that the role lncRNAs play in BC still requires further attention.
Nevertheless, from analyzing the functions of VIM-AS1, SNHG8, and ELDR genes in tumor genesis and development, we believe these three key genes screened are deserving of further research. To conclude, this study enriched the differential expression profile of serum exosomes of BC patients, searched for and identified the target genes of candidate lncRNAs through bioinformatic methods, and predicted their functions and related signaling pathways. This has ultimately provided more reasons for further research on the role and mechanism of lncRNAs in BC. In addition, due to the small sample size of this study, future research on the role of lncRNAs should be expanded further to verify the appropriateness of our lncRNA candidate selection. This could be carried out by using real-time fluorescence quantitative PCR technology and molecular biology to verify and clarify the biological functions of lncRNAs in the development and occurrence of tumors.
Acknowledgments
Funding: This work was supported by funding from the Joint Construction Project of Medical Science and Technology of Henan Province (grant No. LHGJ20190635).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-3483
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-3483
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-3483). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was undertaken following the World Medical Association (WMA) Declaration of Helsinki (as revised in 2013), the National Health and Family Planning Commission’s Measures for the Ethical Review of Biomedical Research Involving Human Beings, and other relevant laws and regulations. It was approved by the Medical Ethics Committee of Henan Cancer Hospital (V1.0-20191029). All the subjects gave their consent and signed the relevant consent form.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Wang M, Sun X, Wang H, et al. Long non-coding RNAs in non-small cell lung cancer: functions and distinctions from other malignancies. Transl Cancer Res 2019;8:2636-53. [Crossref]
- Liu Y, Sharma S, Watabe K. Roles of lncRNA in breast cancer. Front Biosci (Schol Ed) 2015;7:94-108. [Crossref] [PubMed]
- Dykes IM, Emanueli C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genomics Proteomics Bioinformatics 2017;15:177-86. [Crossref] [PubMed]
- Tomasetti M, Lee W, Santarelli L, et al. Exosome-derived microRNAs in cancer metabolism: possible implications in cancer diagnostics and therapy. Exp Mol Med 2017;49:e285 [Crossref] [PubMed]
- Teng Y, Ren Y, Hu X, et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun 2017;8:14448. [Crossref] [PubMed]
- Peinado H, Lavotshkin S, Lyden D. The secreted factors responsible for pre-metastatic niche formation: old sayings and new thoughts. Semin Cancer Biol 2011;21:139-46. [Crossref] [PubMed]
- Ratajczak J, Wysoczynski M, Hayek F, et al. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia 2006;20:1487-95. [Crossref] [PubMed]
- Mashouri L, Yousefi H, Aref AR, et al. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer 2019;18:75. [Crossref] [PubMed]
- Yamada J, Jinno S. Promotion of synaptogenesis and neural circuit development by exosomes. Ann Transl Med 2019;7:S323. [Crossref] [PubMed]
- Zhang X, Yuan X, Shi H, et al. Exosomes in cancer: small particle, big player. J Hematol Oncol 2015;8:83. [Crossref] [PubMed]
- Wu CY, Du SL, Zhang J, et al. Exosomes and breast cancer: a comprehensive review of novel therapeutic strategies from diagnosis to treatment. Cancer Gene Ther 2017;24:6-12. [Crossref] [PubMed]
- Ewaisha R, Gawryletz CD, Anderson KS. Crucial considerations for pipelines to validate circulating biomarkers for breast cancer. Expert Rev Proteomics 2016;13:201-11. [Crossref] [PubMed]
- Miao Y, Fan R, Chen L, et al. Clinical Significance of Long Non-coding RNA MALAT1 Expression in Tissue and Serum of Breast Cancer. Ann Clin Lab Sci 2016;46:418-24. [PubMed]
- Zhang K, Luo Z, Zhang Y, et al. Circulating lncRNA H19 in plasma as a novel biomarker for breast cancer. Cancer Biomark 2016;17:187-94. [Crossref] [PubMed]
- Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9. [Crossref] [PubMed]
- Zhao R, Zhang Y, Zhang X, et al. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer 2018;17:68. [Crossref] [PubMed]
- Xing F, Liu Y, Wu SY, et al. Loss of XIST in Breast Cancer Activates MSN-c-Met and Reprograms Microglia via Exosomal miRNA to Promote Brain Metastasis. Cancer Res 2018;78:4316-30. [Crossref] [PubMed]
- Tang S, Zheng K, Tang Y, et al. Overexpression of serum exosomal HOTAIR is correlated with poor survival and poor response to chemotherapy in breast cancer patients. J Biosci 2019;44:37. [Crossref] [PubMed]
- Wang YL, Liu LC, Hung Y, et al. Long non-coding RNA HOTAIR in circulatory exosomes is correlated with ErbB2/HER2 positivity in breast cancer. Breast 2019;46:64-9. [Crossref] [PubMed]
- Wang X, Pei X, Guo G, et al. Exosome-mediated transfer of long noncoding RNA H19 induces doxorubicin resistance in breast cancer. J Cell Physiol 2020;235:6896-904. [Crossref] [PubMed]
- Zhong G, Wang K, Li J, et al. Determination of Serum Exosomal H19 as a Noninvasive Biomarker for Breast Cancer Diagnosis. Onco Targets Ther 2020;13:2563-71. [Crossref] [PubMed]
- Boque-Sastre R, Soler M, Oliveira-Mateos C, et al. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc Natl Acad Sci U S A 2015;112:5785-90. [Crossref] [PubMed]
- Rezanejad Bardaji H, Asadi MH, Yaghoobi MM. Long noncoding RNA VIM-AS1 promotes colorectal cancer progression and metastasis by inducing EMT. Eur J Cell Biol 2018;97:279-88. [Crossref] [PubMed]
- Sur S, Nakanishi H, Steele R, et al. Long non-coding RNA ELDR enhances oral cancer growth by promoting ILF3-cyclin E1 signaling. EMBO Rep 2020;21:e51042 [Crossref] [PubMed]