Genomic instability-associated lncRNA signature predicts prognosis and distinct immune landscape in gastric cancer
Introduction
As a heterogeneous disease with a worldwide presence, gastric cancer (GC) is the fifth most common malignant tumor and the third leading cause of cancer-related mortality worldwide (1). Despite efforts in the past decade to improve treatment effects, treatment outcomes for GC remain unsatisfactory. Previous studies have concluded that less than one-third of GC cases worldwide are diagnosed at an early stage (2). Based on data from the National Cancer Database Research Unit of the American College of Surgeons, GC patients in east Asia were diagnosed at a relatively early stage and the survival rate was higher; however, there is still much room for the improvement of treatment outcomes (3). The complex classification of GC shows its strong heterogeneity. Based on phenotype observed under a microscope, GC is categorized to three major histological subtypes based on Lauren’s classification, as follows, intestinal, diffuse, and mixed types (4). Though considered as a relatively old and rough classification method, it is still widely used because of its simplicity and robustness. Currently, the World Health Organization (WHO) classification system is the most widely acknowledged pathological system, which divides GC into four main types, namely, papillary adenocarcinoma, tubular adenocarcinoma, mucinous adenocarcinoma, and poorly cohesive carcinoma (5). The last decade has seen the establishment of different molecular classifications of GC. The Cancer Genome Atlas (TCGA) project system, which has attracted the most attention, divides GC into four subtypes, namely, Epstein-Barr virus (EBV)-positive, microsatellite instability (MSI), genomically stable (GS), and chromosomal instability (CIN) GC (6).
As one of the most important signatures of cancer, genomic instability is characterized by the accumulation of somatic mutations. Carcinogenesis tends to result from increasing numbers of somatic mutations, especially driver mutations that affect multiple aspects of tumor growth (7). Tumor mutation burden (TMB) is defined as the total number of somatic mutations present in a single tumor sample. The TMB provides a quantitative description of somatic mutation events in each sample. Previous studies have reported that somatic mutations in tumor DNA can trigger the development of neoantigens, which can be recognized and targeted by the immune system (8,9). The extent of TMB is largely dependent on the tumor type, with melanoma and non-small cell lung cancers (NSCLCs) being associated with the highest TMB and leukemias and pediatric tumors being associated with the lowest TMB. The extent of TMB has been shown to be highly consistent with immune checkpoint blocker (ICB) activity in clinical experiments (10). There is accumulating evidence that a higher TMB is an indicator of a better response to ICB treatment (1-3). A relationship between GCs and genomic instability has been previously reported (4,5). However, it remains unclear how GC is affected by TMB.
The TMB can be affected at multiple biological levels. Defined as transcripts that are longer than 200 nt, long non-coding RNAs (lncRNAs) have been identified as crucial factors affecting tumor behavior. Aberrant expression of lncRNAs has been shown to exert a profound impact on the biological behaviors of cancer such as proliferation, progression, and metastasis (6). It has been found that lncRNAs associated with MSI influence the prognosis of GCs (7); however, the mechanism remains unknown. As one of the members of the ubiquitin-like and ubiquitin-associated (UBL-UBA) protein family, UBQLN4 plays an important role in sustaining genomic stability by affecting nucleotide excision repair. Recent studies have revealed that the mutation of UBQLN4 could reflect genomic instability in cancers, thus affecting prognosis and the chemotherapy response (8). In this study, we established a six-lncRNA-based signature by comparing high and low TMB groups using data obtained from TCGA. The signature was further analyzed to assess tumor immune microenvironment variations. We assessed the predictive value of the signature for sensitivity to chemotherapy and immunotherapy. This study not only provides an explanation for why GC patients with high TMB experience better survival, but also provides clues for predicting the prognosis and drug sensitivity of GC patients. We present the following article in accordance with the STARD reporting checklist (available at https://dx.doi.org/10.21037/atm-21-3569).
Methods
Data source
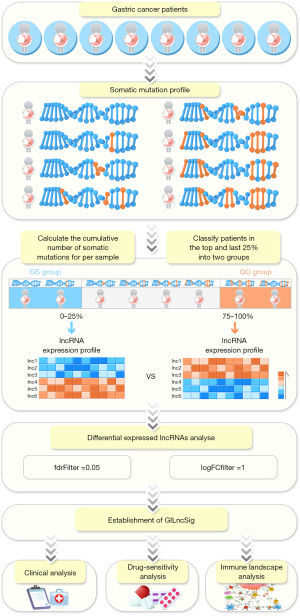
The RNA-seq, clinical, and single nucleotide variant (SNV) data from patients with GC were derived from the TCGA Data Portal (https://portal.gdc.cancer.gov/, 10 December 2020). Integral lncRNA and messenger RNA (mRNA) expression profiles, somatic mutation data, common clinicopathological characteristics and survival data of 375 patients were used for signature establishment. We divided GC patients with follow-up data (time of follow-up =0 were excluded) into two groups, namely, the training cohort and the testing cohort, as previously reported (9). A flowchart of this study is presented in Figure 1. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Identification of genomic instability associated lncRNAs
The RNA sequencing and SNV data from patients with GC from TCGA datasets were used to identify genomic instability-associated lncRNAs. First, SNV data from all the patients were rearranged in order of decreasing number of cumulative somatic mutations. The top quarter of patients were defined as the genomically unstable (GU) group (n=91), while the bottom quarter were defined as the GS group (n=90). The “Limma” R package was applied to identify the differentially expressed lncRNAs under the thresholds of |log2 FC| >1 and P value <0.05. The lncRNAs that met the criteria were regarded as genomic instability-concerned.
Functional enrichment analysis
The paired expression profiles of lncRNAs and mRNAs were matched based on the Pearson correlation algorithm. The mRNAs with higher correlations (top 10) were regarded as the co-expressed partners of their corresponding lncRNAs. The selected mRNAs were subjected to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis.
Construction and evaluation of genomic instability lncRNA-based gene signature
By using Ward’s linkage and Euclidean distance methods, hierarchical cluster analyses were performed to separate GC patients into two groups, namely, the GU-like group (n=318) and the GS-like group (n=57). Cox regression analysis was conducted to identify genomic instability-associated lncRNAs with prognostic value. The selected lncRNAs were used to construct a genomic instability-associated lncRNA signature (GILncSig) scoring system for survival prediction with the following formula:
In Eq. [1], lncRNAi represents the prognosis-related lncRNAs derived from Cox regression analysis; expr(lncRNAi) indicates the expression of lncRNAi; and coef(lncRNAi) is the predictive value of lncRNAi for GILncSig-based risk scores, which were obtained from the multivariate Cox analysis. Patients in the training cohort were divided into high-risk group and low-risk group based on their risk scores.
Kaplan-Meier curves were applied to calculate the survival difference between the high- and low-risk groups with a threshold of P value ≤5%. The newly established GILncSig was further tested by Cox regression analysis to assess whether it is a better independent prognostic factor for GC than clinical features such as tumor, node, metastasis (TNM) stage. A receiver operating characteristic (ROC) curve was also drawn to validate the performance of the signature in predicting prognosis.
Exploration of immune landscapes based on different risk stratifications
Based on the transcriptional data derived from TCGA dataset, single-sample Gene Set Enrichment Analysis (ssGSEA) was performed with “ssGSEA” in R software (3.6.1) to elucidate the immune spectra. The “GSVA” package in R software 3.6.1 (http://www.R-project.org/) was also utilized for the presentation of ssGSEA results. At the same time, the ESTIMATE algorithm was utilized by using the “estimate” package in R (3.6.1) to obtain the immune score, stromal score, and ESTIMATE score (the sum of the previous two data points) for each sample from GC patients obtained from TCGA. To determine the relationship between the risk score and the tumor immune microenvironment, currently well-known methods, including TIMER, XCELL, QUANTISEQ, MCP-counter, EPIC, and CIBERSORT, were applied to calculate the proportions of infiltrating immune cells. Spearman correlation analysis was further conducted to analyze the correlations between the GILncSig and the immune cells. The correlation coefficients of the results are shown in a lollipop diagram under the threshold of R>0.1 and P<0.05.
Evaluation of the response of chemotherapy and immunotherapy based on different risk stratifications
To explore the sensitivity of each GC patient from TCGA to different chemotherapeutic agents, the “pRRophetic” package in R 3.6.1 was applied to predict the half maximal inhibitory concentration (IC50) of GC-related chemotherapy drugs, and the high- and low-risk groups were compared. This algorithm was published previously and has been widely used in several studies (10,11). The online tool Tumor Immune Dysfunction and Exclusion (TIDE) (http://tide.dfci.harvard.edu) was used to predict the TIDE score, which has been shown to be positively correlated with anti-PD1 and anti-CTLA4 immunotherapy responses. The immunophenoscore (IPS) refers to the four parts [immunosuppressive cells, effector cells, major histocompatibility complex (MHC) molecules and immunomodulators] determining immunogenicity which is calculated without bias using machine learning methods. The IPS (range: 0 to 10) is calculated based on the gene expression in representative cell types. The IPS results of stomach adenocarcinoma (STAD) patients were downloaded from The Cancer Immunome Atlas (TCIA) (https://tcia.at/home).
Statistical analysis
The statistical analyses were all generated by R-3.6.1 in this study. For quantitative data, statistical significance for nonnormally distributed variables were estimated by the Wilcoxon rank-sum test, and normally distributed variables were analyzed by Student’s t-tests. Kruskal-Wallis tests and one-way analysis of variance were used as nonparametric and parametric methods for comparisons of more than two groups, respectively. Spearman method was applied for correlation analysis between two continuous variables. Contingency tables were analyzed by two-sided Fisher exact tests. Kaplan-Meier survival analysis and the Cox proportional hazards model were used to analyze the association between the two risk stratifications with the R package ‘Survminer’. The prognosis classification performance of the SRLncSig score model was assessed by the ROC curve, and the area under the curve (AUC) were calculated using ‘timeROC’ package.
Results
Identification of genomic instability-related lncRNAs in TCGA datasets
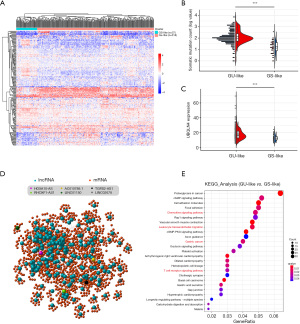
Genomic instability-related lncRNAs were identified based on differential expression analysis between the groups with high and low numbers of cumulative somatic mutation. The top one-quarter of patients were defined as the GU group (n=91), while the bottom one-quarter were defined as the GS group (n=90). The “Limma” R package was applied to identify the differentially expressed lncRNAs under the thresholds of |log2 FC| >1 and P value <0.05. There were 53 upregulated and 144 downregulated lncRNAs that met the criteria (Table S1). The top 40 genes ranked by |log2 FC| are presented in Figure S1A. Based on the 197 differentially expressed lncRNAs that were discovered, hierarchical cluster analyses were performed to separate the GC patients into two groups, namely, the GU-like group (n=318) and GS-like group (n=57) (Figure 2A). The somatic mutation counts are presented in Figure 2B. The expression level of UBQLN4 was also higher in the GU-like group than in the GS-like group (Figure 2C).
Functional enrichment analysis
Based on the method presented above, we established a lncRNA-mRNA network based on correlation analysis. The red nodes indicate lncRNAs, while the blue nodes represent mRNAs (Figure 2D). The details are all listed in https://cdn.amegroups.cn/static/public/atm-21-3569-1.xlsx. The lncRNA-correlated mRNAs were further used to conduct KEGG pathway analysis. The results revealed that immune-related pathways, such as the chemokine signaling pathway, T cell receptor signaling pathway, and leukocyte transendothelial migration pathway, were enriched. The GC-related pathways, such as gastric acid secretion, GC and focal adhesion, were also enriched. These results indicate that genomic instability-related lncRNAs are highly correlated with GC development and that immune landscape changes could play a role in the disease (Figure 2E).
Establishment and validation of the robustness of 6-lncRNA based GILncSig
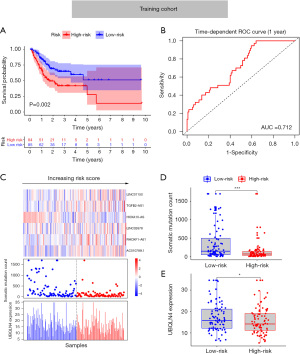
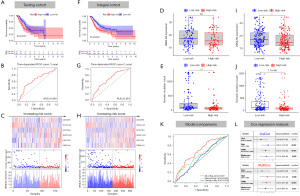
To improve the prognostic predictive value of the genome instability-associated lncRNAs, TCGA GC patients were randomly divided into two cohorts, namely, a training cohort (n=169) and a testing cohort (n=168). To identify the lncRNAs prognostic value, the training cohort was used to establish a signature by conducting univariate Cox regression analysis of the 197 genomic instability-related lncRNAs. Of all the 197 genomic instability-related lncRNAs, 8 lncRNAs were identified to be prognosis-related (P<0.05; Figure S1B). Multivariate Cox regression analysis of the eight selected lncRNAs was carried out, and 6 lncRNAs with independent prognostic value, namely, LINC02678, HOXA10-AS, RHOXF1-AS1, AC010789.1, LINC01150, and TGFB2-AS1, were identified. We established GILncSig based on the values of coefficients from multivariate Cox analysis and the expression level of the six lncRNAs as follows: GILncSig risk score = (0.1485 × expression level of LINC02678) + (–0.5348 × expression level of HOXA10-AS) + (0.0547 × expression level of RHOXF1-AS1) + (0.245 × expression level of AC010789.1) + (0.7888 × expression level of LINC01150) + (0.0917 × expression level of TGFB2-AS1). The risk score of each TCGA GC sample was obtained based on the formula. First, the risk score was used to separate the training cohort into low and high-risk groups. A Kaplan-Meier curve was applied, and it was found that the low-risk group had better survival than the high-risk group (Figure 3A). An ROC curve was also drawn to assess the predictive value of the signature for survival (AUC =0.712) (Figure 3B). The median of the risk score was set as the cutoff value (Figure 3C) and the low-risk group presented with a larger number of somatic mutations (Figure 3D) and a higher expression level of UBQLN4 than the high-risk group (Figure 3E). The established GILncSig was further applied to test it’s prognosis-predictive value in the testing cohort. The results were highly consistent except for the insignificant expression levels of UBQLN4 between two risk groups (Figure 4A-4E). Finally, the integral cohort was examined by the GILncSig. The results were also highly consistent (Figure 4F-4J). Compared with other GC-related lncRNA signatures (WangLncSig’ AUC =0.589 and ZhangLncSig’ AUC =0.536), GILncSig performed better in prognosis prediction in TCGA STAD datasets (GILncSig’ AUC =0.681) (Figure 4K) (12,13). Moreover, GILncSig was identified as an independent prognostic factor based on univariate and multivariate Cox regression analysis (Figure 4L). The clinical baseline data of the two cohorts were also compared, and no statistically significant differences were found (Table S1). The predictive value of GILncSig was further tested based on different clinical characteristics and other GC-related lncRNA signatures. The results indicated that GILncSig is still a good prognostic predictor regardless of age, gender, tumor grade, and TNM stage (Figure 5A-5H).
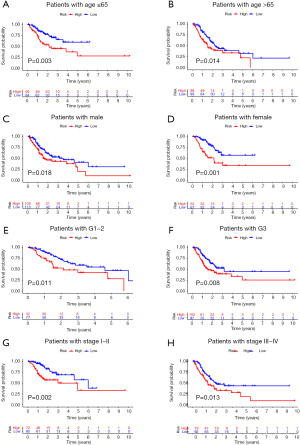
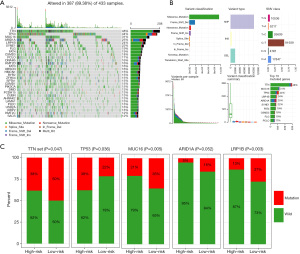
Correlations between GILncSig and SNVs
Somatic mutations in MUC16 and TNN have been reported as correlated with better survival in GC (14). The SNV datasets of TCGA STAD projects were acquired and the top 30 frequently mutated genes were presented by waterfall maps (Figure 6A). The details of the mutations were also summarized (Figure 6B). As for the GILncSig, the high-risk group presented with higher mutation rates for most of the genes. The top 5 genes were enrolled for the comparison between two groups. The results indicated that most of the genes harbored higher mutation rates in the high-risk group (Figure 6C).
Differences in tumor immune microenvironment landscape based on risk stratification
The ESTIMATE algorithm was used to determine ESTIMATE scores, immune scores, and stromal scores, and the two risk groups were compared (Figure 7A). The results indicated that ESTIMATE scores, immune scores, and stromal scores were significantly higher in the high-risk cohort than the low-risk cohort (Figure 7B). To further evaluate the hidden functional components, the mainstream software for qualifying the infiltrative immune cells were applied. Immune cells generally considered to have anti-tumor function for higher antigen-processing events (T cells CD4+ memory cells, CD8+ T cells, and T cells CD8+ effector memory cells) were higher in the low-risk group while cells with immune regularity function (M2 macrophages and cancer-associated fibroblasts) were higher in the high-risk group (Figure 7C).
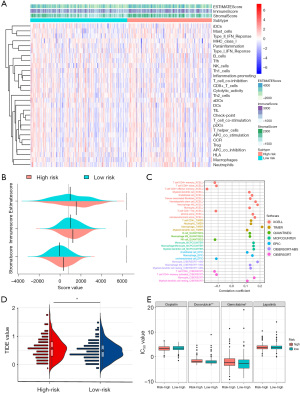
Distinct response to immunotherapy and chemotherapy in the high- and low-risk groups
The online tool TIDE was applied to investigate the predictive value of immune checkpoint inhibitors based on the newly established signature. The results revealed that the TIDE value was significantly higher in the high-risk group than the low-risk group, which indicates a favorable immunotherapy response for the high-risk group (Figure 7D). We predicted the IC50 of four GC-concerning chemotherapy drugs included in the “pRRophetic” package (cisplatin, doxorubicin, gemcitabine, and lapatinib) in samples from high- and low-risk patients with the pRRophetic algorithm (Figure 7E). The results revealed that the IC50 values of doxorubicin and gemcitabine were higher in the high-risk group than the low-risk group. It can be inferred that the high-risk group is more likely to benefit from immune checkpoint therapy and that risk stratification can be a predictor of the sensitivity of GC patients to different drugs.
Discussion
The past few years have witnessed the explosion of ICB treatment regardless of clinical experiments or basic studies (3). There have been various challenges for GC-related studies due to the negative outcomes obtained in clinical trials. However, numerous studies have indicated that genomic instability is a valuable predictor of immunotherapy and chemotherapy responses (15). Genomic instability has been revealed to be a unique feature of cancers and to influence the tumor immune microenvironment in GC (16,17). Genomic instability may play a crucial role in cancer progression and recurrence and may affect the prognosis of cancer in multiple ways (9,18). For example, low-grade gliomas (a tumor type with relatively low genomic instability), chemotherapy with temozolomide was associated with high rates of tumor progression. Meanwhile, the study also discovered that at least half of the mutations in the initial tumor were undetected at recurrence including driver mutations in TP53, ATRX, SMARCA4, and BRAF; this suggests that recurrent tumors are often seeded by cells derived from the initial tumor at a very early stage of their evolution (19). Previous studies have suggested that many somatic gene mutations could predict better survival in GC patients (14,20,21). Of all the reported genes, MUC16 has attracted the most attention because of the highly consistent findings in both Eastern and Western countries (20). Our results confirm the results of previous studies, however from another perspective. The five most frequently mutated genes were also extracted to make a comparison based on the established GILncSig. The result indicated the effectiveness of GILncSig for genomic state prediction. The genomic instability reflected by somatic mutation counts was higher in low-risk patients who presented with better survival than high-risk patients. The results are controversial because most of the reported mutated genes in other types of tumors have been shown to indicate a worse prognosis. The variation in immune landscape between the two risk groups may explain this finding. As presented in the results, the proportions of immune cells generally considered to have antitumor function for higher antigen-processing events (memory activated CD4 T cells and follicular helper T cells) were higher in the low-risk group than the high-risk group, while the proportions of cells with immune regularity function (regulatory T cells and M2 macrophages) were higher in the high-risk group. The results can be explained by the fact that the immune system is triggered by accumulating somatic mutations. However, direct evidence for immune landscape changes is still limited. To further explore the clinical application value of GILncSig, we used two ICB therapy-related algorithms to estimate the predictive value of this signature for immunotherapy. Both algorithms revealed that the high-risk group may be more likely to benefit from ICB therapy. The results may indicate that GILncSig has potential for use in guiding ICB therapies.
Multiple research fields have linked lncRNAs and GC. The novel lncRNA, GClnc1 could play a role in recruiting both WDR5 and the KAT2A complex to affect the transcription of target genes and promote the carcinogenesis of GC (22). Circulating exosomal lncRNA-GC1 was discovered to have the potential to be developed into a noninvasive indicator for the early detection of GCs (23). Unfortunately, the study was established based on a single center cohort, more validations cohort were needed to confirm the effeteness of this method. Autophagy can be regulated by lncRNA-MALAT1 via miR-23b-3p sequestration in GC, and lncRNA-MALAT1 was also revealed to be associated with chemoresistance (24). Although the related studies are too numerous to list, few have tried to explore the potential link between genomic instability-related lncRNAs and GC. A total of 16 lncRNA-based signatures were identified as predictors to classify GC patients’ MSI status, and the signature was also discovered to have the potential to distinguish patients with good survival from those with poor survival (7). Most of the lncRNAs included in GILncSig were found to be associated with multiple cancers other than GC. The lncRNA, AC010789.1, was reported to promote the progression of colorectal cancer by targeting the microRNA-432-3p/ZEB1 axis and activating the Wnt/β-catenin signaling pathway (25). It has been reported that HOX-lncRNAs have the capacity to influence the carcinogenesis of lung cancer (26). Transforming growth factor (TGF)-β-induced TGFB2-AS1 lncRNA was found to have an inhibitory impact on TGF-β/BMP signaling (27). However, more investigations are needed to explore the biological function of the six lncRNAs identified in this study.
As an important protein that is involved in DNA double-strand break repair, UBQLN4 has attracted increasing attention in recent years. Regardless of the mutation state or expression level, UBQLN4 was revealed to be an excellent indicator of genomic state (8). Recently, GC-related studies on UBQLN4 have emerged. In GCs, UBQLN4 may play a protective role to prevent the progression of cancer cells by affecting the stability of P21 in a p53-independent or p53-dependent manner (28). Moreover, UBQLN4 has also been revealed to have the potential to affect cell cycle arrest and suppress the progression of GC. The results of this study are consistent with ours in terms of UBQLN4’s tumor suppressive role in GCs.
We were driven to further explore the effect of genomic instability on the immune spectrum for two reasons. First, functional enrichment analysis identified pathways previously found to be associated with GC and pathways that play crucial roles in the immune response. Second, the low-risk group, which presented with better survival, had more severe genomic instability. As mentioned previously, somatic mutations in genes, especially driver mutations, can induce the development of neoantigens, which can be recognized by the adaptive immune system to develop a specific immune reaction. Functional mutations can be transcribed and translated into neoantigen-containing peptides and presented by MHC molecules on the cell surface. However, only a small portion of mutations lead to the development of neoantigen-containing peptides that can be properly processed and presented by MHC complexes. Moreover, even fewer of these peptides can be recognized properly by T cells (29). Although the theory can partially explain why the low-risk group had a higher TMB than the high-risk group, it remains unclear why this phenomenon is completely different between GC and breast cancer. The potential explanations might be attributed to the accumulations of gene mutations. Higher level of TMB had been revealed as important trigger of anti-cancer response and thus leaded to better prognosis of low-risk group. However, the hypothesis remained to be proved (30).
Besides lncRNAs, other non-coding RNAs also played a role in genomic instability. For examples, genomic instability-derived plasma extracellular vesicle-microRNA signature was established and perform as a minimally invasive predictor of risk and unfavorable prognosis in breast cancer. The correlations between microRNA and genomic instability characteristics were also revealed (30). Circular RNAs were also identified as important factors affecting the age-related genomic regulation (31). As for the therapeutic potential of genomic instability, relevant studies were emerging in recent years. Genomic instability and CIN are nearly ubiquitous feature of human cancers, and a therapy that can exploit the fitness tradeoffs associated with CIN under the condition of the normal function of healthy diploid cells would represent a critical step forward in precision oncology. In summary, these studies implicated that spindle assembly checkpoint (SAC) protein KIF18A may be a therapeutic target that is specifically required in cells characterized by CIN or other related ploidy abnormalities (32).
Although our study has provided fresh insight into the impact of genome instability on the prognosis of GC patients, limitations still exist. Considering that GILncSig was established based on the TCGA cohort, which consists of whole exon sequencing and whole transcriptional sequencing data, it is difficult to validate the signature using other data. The efficiency of GILncSig needs further validation. In addition, GILncSig was identified based on bioinformatics. The results should be further confirmed by biological experiments.
Conclusions
This work proposed a computational frame derived from the mutator hypothesis to identify genome instability-associated lncRNAs. The combination analysis of lncRNA expression profiles with somatic mutation profiles and clinical information of GC patients in TCGA STAD datasets helped us to identify a genome instability-derived lncRNA signature as an independent prognostic marker to stratify risk subgroups. The prior KEGG analysis based on the mRNAs correlated with genome instability-associated lncRNAs further drove us to study the immune landscape changes based on different risk subgroups. In addition, the IPS and TIDE value and estimated IC50 for chemotherapy drugs were also calculated for comparison of different risk subgroups. The results also revealed the predictive value of therapy response for the GILncSig. Through further prospective validation, the GILncSig may have important implications for genome instability and customized decision-making in GC patients.
Acknowledgments
Funding: This work was supported by grants from the National Natural Science Foundation of China (81872425, 81972228) and Shanghai Pu Jiang Talents plan (2019PJD005).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-3569
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-3569). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jardim DL, Goodman A, de Melo Gagliato D, et al. The challenges of tumor mutational burden as an immunotherapy biomarker. Cancer Cell 2021;39:154-73. [Crossref] [PubMed]
- Gandara DR, Paul SM, Kowanetz M, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med 2018;24:1441-8. [Crossref] [PubMed]
- Chan TA, Yarchoan M, Jaffee E, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol 2019;30:44-56. [Crossref] [PubMed]
- Ottini L, Falchetti M, Lupi R, et al. Patterns of genomic instability in gastric cancer: clinical implications and perspectives. Ann Oncol 2006;17:vii97-102. [Crossref] [PubMed]
- Suzuki K, Ohnami S, Tanabe C, et al. The genomic damage estimated by arbitrarily primed PCR DNA fingerprinting is useful for the prognosis of gastric cancer. Gastroenterology 2003;125:1330-40. [Crossref] [PubMed]
- Sanchez Calle A, Kawamura Y, Yamamoto Y, et al. Emerging roles of long non-coding RNA in cancer. Cancer Sci 2018;109:2093-100. [Crossref] [PubMed]
- Chen T, Zhang C, Liu Y, et al. A gastric cancer LncRNAs model for MSI and survival prediction based on support vector machine. BMC Genomics 2019;20:846. [Crossref] [PubMed]
- Jachimowicz RD, Beleggia F, Isensee J, et al. UBQLN4 represses homologous recombination and is overexpressed in aggressive tumors. Cell 2019;176:505-519.e22. [Crossref] [PubMed]
- Bao S, Zhao H, Yuan J, et al. Computational identification of mutator-derived lncRNA signatures of genome instability for improving the clinical outcome of cancers: a case study in breast cancer. Brief Bioinform 2020;21:1742-55. [Crossref] [PubMed]
- Geeleher P, Cox NJ, Huang RS. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol 2014;15:R47. [Crossref] [PubMed]
- Song C, Guo Z, Yu D, et al. A prognostic nomogram combining immune-related gene signature and clinical factors predicts survival in patients with lung adenocarcinoma. Front Oncol 2020;10:1300. [Crossref] [PubMed]
- Wang Y, Zhang H, Wang J. Discovery of a novel three-long non-coding RNA signature for predicting the prognosis of patients with gastric cancer. J Gastrointest Oncol 2020;11:760-9. [Crossref] [PubMed]
- Zhang X, Zhang W, Jiang Y, et al. Identification of functional lncRNAs in gastric cancer by integrative analysis of GEO and TCGA data. J Cell Biochem 2019;120:17898-911. [Crossref] [PubMed]
- Yang Y, Zhang J, Chen Y, et al. MUC4, MUC16, and TTN genes mutation correlated with prognosis, and predicted tumor mutation burden and immunotherapy efficacy in gastric cancer and pan-cancer. Clin Transl Med 2020;10:e155 [Crossref] [PubMed]
- Pietrantonio F, Miceli R, Raimondi A, et al. Individual patient data meta-analysis of the value of microsatellite instability as a biomarker in gastric cancer. J Clin Oncol 2019;37:3392-400. [Crossref] [PubMed]
- Kim J, Kim B, Kang SY, et al. Tumor mutational burden determined by panel sequencing predicts survival after immunotherapy in patients with advanced gastric cancer. Front Oncol 2020;10:314. [Crossref] [PubMed]
- Parikh AR, He Y, Hong TS, et al. Analysis of DNA damage response gene alterations and tumor mutational burden across 17,486 tubular gastrointestinal carcinomas: implications for therapy. Oncologist 2019;24:1340-7. [Crossref] [PubMed]
- Mettu RK, Wan YW, Habermann JK, et al. A 12-gene genomic instability signature predicts clinical outcomes in multiple cancer types. Int J Biol Markers 2010;25:219-28. [Crossref] [PubMed]
- Johnson BE, Mazor T, Hong C, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 2014;343:189-93. [Crossref] [PubMed]
- Li X, Pasche B, Zhang W, et al. Association of MUC16 mutation with tumor mutation load and outcomes in patients with gastric cancer. JAMA Oncol 2018;4:1691-8. [Crossref] [PubMed]
- Dong MM, Peng SJ, Yuan YN, et al. LncRNA TTN-AS1 contributes to gastric cancer progression by acting as a competing endogenous RNA of miR-376b-3p. Neoplasma 2019;66:564-75. [Crossref] [PubMed]
- Sun TT, He J, Liang Q, et al. LncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discov 2016;6:784-801. [Crossref] [PubMed]
- Guo X, Lv X, Ru Y, et al. Circulating exosomal gastric cancer-associated long noncoding RNA1 as a biomarker for early detection and monitoring progression of gastric cancer: a multiphase study. JAMA Surg 2020;155:572-9. [Crossref] [PubMed]
- YiRen H. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer 2017;16:174. [Crossref] [PubMed]
- Duan W, Kong X, Li J, et al. LncRNA AC010789.1 promotes colorectal cancer progression by targeting microRNA-432-3p/ZEB1 axis and the Wnt/β-catenin signaling pathway. Front Cell Dev Biol 2020;8:565355 [Crossref] [PubMed]
- Li L, Wang Y, Song G, et al. HOX cluster-embedded antisense long non-coding RNAs in lung cancer. Cancer Lett 2019;450:14-21. [Crossref] [PubMed]
- Papoutsoglou P, Tsubakihara Y, Caja L, et al. The TGFB2-AS1 lncRNA regulates TGF-β signaling by modulating corepressor activity. Cell Rep 2019;28:3182-98.e11. [Crossref] [PubMed]
- Huang S, Li Y, Yuan X, et al. The UbL-UBA Ubiquilin4 protein functions as a tumor suppressor in gastric cancer by p53-dependent and p53-independent regulation of p21. Cell Death Differ 2019;26:516-30. [Crossref] [PubMed]
- Coulie PG, Van den Eynde BJ, van der Bruggen P, et al. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer 2014;14:135-46. [Crossref] [PubMed]
- Bao S, Hu T, Liu J, et al. Genomic instability-derived plasma extracellular vesicle-microRNA signature as a minimally invasive predictor of risk and unfavorable prognosis in breast cancer. J Nanobiotechnology 2021;19:22. [Crossref] [PubMed]
- Bravo JI, Nozownik S, Danthi PS, et al. Transposable elements, circular RNAs and mitochondrial transcription in age-related genomic regulation. Development 2020;147:dev175786 [Crossref] [PubMed]
- Bielski CM, Taylor BS. Homing in on genomic instability as a therapeutic target in cancer. Nat Commun 2021;12:3663. [Crossref] [PubMed]
(English Language Editor: J. Jones)