
SLC7A5 serves as a prognostic factor of breast cancer and promotes cell proliferation through activating AKT/mTORC1 signaling pathway
Introduction
According to the 2020 global Cancer Statistics, breast cancer (BC) is the most commonly diagnosed malignancy in females worldwide (1). BC is a typical heterogeneous cancer, which displays significant diversity depending on the particular molecular signature (2), such as estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). Furthermore, based on the above molecular signatures, BC can be divided into 4 subtypes: the luminal A, luminal B, HER2, and basal-like subtypes (3). One avenue of cancer research involves examining the molecular profiles of the cancer cells and their possible relationship with the clinical outcomes of the disease process.
The growth of tumor cells relies on amino acids (AAs), as they enhance mammalian targets of rapamycin complex 1 (mTORC1) via nutritional signaling pathways for regulation of protein synthesis and cancer survival (4,5). Studies have suggested that mammalian target of rapamycin complex 1(mTORC1) plays a role in reprogramming tumor cell metabolism, specifically, in the direct and indirect regulation of AA transport (6).
As a vital member of AA transporters, solute carrier family 7 member 5 (SLC7A5) is critical for the sodium-independent transport of large neutral AAs across the membrane (7). This biological process is essential for the rapid growth and proliferation of tumor cells (8). Altered regulation of SLC7A5 has been found in different cancer types, such as ovarian cancer and non-small cell lung cancer (NSCLC) (9-11).
In our research, we analyzed the gene copy number, messenger RNA (mRNA) expression, and the protein expression of SLC7A5 to examine its prognostic relevance in BC patients in various molecular subtypes. We found that upregulated SLC7A5 expression in MCF-7 cells mediates the AKT/mTORC1 signaling pathway, which is critical for the regulation of tumor cell proliferation, indicating that SLC7A5 might be a potential prognostic parameter and therapeutic target of BC. We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/atm-21-2247).
Methods
BC dataset source and preprocessing
We used Oncomine, an online microarray database (http://www.oncomine.org) that includes 970 datasets and 84,529 samples to determine SLC7A5 mRNA expression in various types of cancer tissues and normal tissues (12). Difference of transcriptional expression was compared by Student’s t-test, and the cutoffs were defined as follows: P level <0.01, fold change >1.5, top gene rank =10%, and a data type of mRNA. Additionally, the expression levels of SLC7A5 in 32 cancer types were analyzed and compared based on the mRNA sequencing (RNA-seq) data of The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/) from the Tumor Immune Estimation Resource (TIMER) database (13). Clinical characteristics such as age, race, tumor-node-metastasis (TNM) stages, and cancer subtypes were obtained with regard to their corresponding clinical patterns. Using the “pROC” package in R (R Foundation of Statistical Computing), we tested the specificity and sensitivity of predictive accuracy of SLC7A5 with receiver operating characteristic (ROC) analysis and area under the curve (AUC).
Genetic variation of SLC7A5 in BC
The invasive breast carcinoma dataset was downloaded from TCGA on the cBioPortal website (14,15). The levels of mRNA expression were retrieved as RNA-seq by expectation-maximization (RSEM; batch-normalized from Illumina HiSeq_RNASeqV2). For missing values, the gene patterns and clinical signatures were filtered.
Survival analyses
We made Kaplan-Meier plots and used log-rank tests to find the significant relevance between SLC7A5 expression and prognosis (16). We evaluated the correlation of SLC7A5 with overall survival (OS) and relapse-free survival (RFS), and calculated hazard ratios (HRs) with 95% confidence intervals (CIs) using uni- and multivariable Cox regression models. Clinical parameters associated with survival were incorporated into the final multivariate prognostic analysis.
Cell lines and cell culture
The human BC cell lines MCF-7 were obtained from the Women’s Hospital of Nanjing Medical University, Nanjing Maternity and Child Health Care Hospital. The cell lines were freshly recovered from a liquid nitrogen. The BC cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, CA, USA) together with the 10% fetal bovine serum (FBS, Gibco, Cappinas, Brazil) in a humidified incubator at a temperature of 37 °C with 5% CO2.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Women’s Hospital of Nanjing Medical University (No.: KY2021054) and informed consent was taken from all individual participants.
Cell transfection
SLC7A5 overexpression lentiviruses were constructed with GV208 by GeneChem Company (Shanghai, China). MCF-7 cells were seeded into 6-well plates, and lentiviral transfection with experimental constructs or respective controls (1×109 TU/mL) was carried out at 60% confluency, following the manufacturer’s protocol. After 48–72 hours, infection efficiency was detected and SLC7A5 expression levels were verified by Western blotting.
Histopathology
The breast cancer tissues were preserved in 10% formalin for histopathology (Solarbio, Beijing, China) and embedded in paraffin and cut into 50-mm thick cubes. The tissues were then stained with 3,3'-diaminobenzidine (DAB) and subjected to immunohistochemical (IHC) staining with standard methods. All figures of pathological changes were taken using an Olympus BX-145 microscope (Olympus, Japan).
Colony formation assay
Six-well plates were covered with a 0.6% agar layer in a medium with FBS (20%). The cells were prepared in 0.3% agar and seeded in 3 parts. After incubations of the plates at a temperature of 37 °C for 2 weeks, we counted the number of colonies.
Cell cycle analysis
Tumor cells were incubated with 10 or 20 µmol/L of naringin for 24 hours and fixed in 75% ethanol at 4 °C overnight for the cell cycle analysis. After resuspension in 10 µg/mL propidium iodide, cell cycle stages were determined using a Coulter Epics XL Flow Cytometry System (Beckman Coulter, Brea, CA, USA).
RT-PCR analysis
TRIzol reagent (Invitrogen, USA) was used to extract the total RNA, and 1 ug of total RNA was then reverse transcribed. The ABI PRISM 7900 sequence detection system (Applied Biosystems, Foster City, CA, USA) was used to perform RT-PCR. We then conducted PCR amplification in a reaction system containing complement DNA (cDNA) using the forward and reverse primers, SYBR Green qPCR Supermix (Thermo Fisher Scientific, USA), and distilled water. The PCR procedure was carried out for 40 cycles at the temperature of 95 °C for 10 seconds and then at a temperature of 60 °C for 30 seconds. Primer sequences for detection of the mRNA expression were synthesized. We evaluated the relative expression of the amplified RNA samples with the 2−∆∆CT method. All reactions were repeated more than 3 times.
Western blot analysis
We collected whole cell lysates using RIPA lysis buffer (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) and calculated the protein concentrations using a BCA Protein Assay Kit (Beyotime, Jiangsu, China). Then, 4–20% sodium dodecyl sulfate polyacrylamide gel electrophoresis (Sigma-Aldrich St. Louis, MO, USA) protein loading buffer was added to the collected 40µg protein samples and heated in a boiling water bath for 3–5 minutes to fully denature the protein. After the protein sample was cooled to room temperature, it was directly loaded into the sample well of SDS-PAGE gel for electrophoresis, which was followed by protein electrophoretic transfer to polyvinylidene fluoride membranes (Roche, Indianapolis, IN, USA). After this, we blocked the membranes in 5% milk, and then incubated them with primary antibodies (Abcam, Cambridge, MA, USA) to evaluate SLC7A5, p-AKT (Abcam), p-p70-S6K, (Abcam), and p-mTOR (Abcam) expression. Subsequent incubations using the secondary antibody were performed, and GAPDH was used as an internal reference.
Statistical analysis
All clinical parameters are shown with mean ± standard error of measurement (SEM) and calculated using Student’s t-test with a P value <0.5 set as the threshold of significance. Experiments were performed in triplicate. Analyses were conducted with R software (version 3.6.1) and Bioconductor packages.
Results
Upregulation of SLC7A5 expression in BC
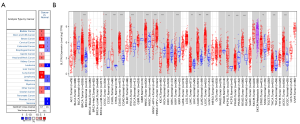
The mRNA of SLC7A5 in different cancers and normal tissues was detected in the Oncomine database. We found an increased level of SLC7A5 in breast, colorectal, lung, lymphoma, and head and neck cancers (Figure 1A). To investigate further SLC7A5 expression in pancreatic cancer, RNA sequencing from TCGA database was analyzed (2). The various expression patterns in cancer and its expression in the adjacent normal tissues are illustrated in Figure 1B. SLC7A5 expression, when compared to its expressions in the normal adjacent tissues, was increased in breast invasive carcinoma, specifically the luminal B, HER2+, and basal-like subtypes, as well as those in colon adenocarcinoma, head and neck squamous carcinoma, lung adenocarcinoma, lung squamous cell carcinoma, rectum adenocarcinoma, and uterine corpus endometrial carcinoma.
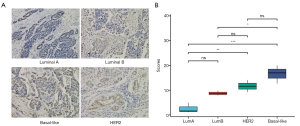
We found that high or low expression level of SLC7A5 protein was found predominantly in invasive BC cells’ membrane (Figure 2A). We used polyclonal SLC7A5 antibody (1:200, Proteintech, USA). The tissue sections were examined by two experienced pathologists using a double-blind method. The difference of positive cells and the intensity of cell staining were considered to score the results of immunohistochemistry Based on the percentages of positive staining cells, the value obtained by multiplying the 2 scores was the final score for each specimen. We also found the differential expression among the four Intrinsic molecular subtypes (PAM50) of breast cancer subtypes (Figure 2B).
Genetic alterations of SLC7A5 in BC
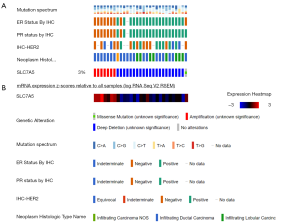
Next, we illustrated the genetic alterations distribution with clinical annotation of SLC7A5 to identify genetic aberrations across the TCGA-BRCA cohort using the cBioPortal OncoPrint module (Figure 3A). According to the cBioPortal definitions, genetic alterations covered the genomic alterations of number aberrations (CNAs) or mutations. In total, 3% of BRCA samples (29/963) harbored at least 1 alteration event within 0.75% and deep deletions in 2.25% of cases in TCGA datasets. The incidence of SLC7A5 point mutations was very limited. We also found that amplification of CNAs was related to high mRNA expression while deep deletion of CNAs was associated with low mRNA expression. This analysis was based on the RNA-Seq by Expectation-Maximization values. Furthermore, the amplificated CNAs were related to ER-/PR-/HER2-negative status and infiltrating ductal carcinoma, while the deep deleted CNAs were associated with ER-/PR-/HER2-positive status and infiltrating lobular carcinoma (Figure 3B).
Correlation between SLC7A5 expression with clinicopathological parameters
In the 1,084 BC patients, 541 patients expressed a low level of SLC7A5 and 542 expressed a high level of SLC7A5. There were significant differences in race, T stage, pathologic stage, histological type, ER status, PR status, and PAM50 subtypes among the 2 subgroups in Table 1 (P value >0.05).

Full table
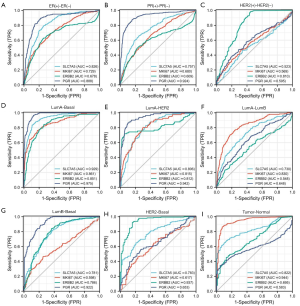
We used the ROC curves to determine the SLC7A5 performance in predicting the outcome of BC subtypes compared with other classical markers (Figure 4). In predicting the outcome of normal or tumor tissues, ER status, and PR status, the predictive ability of the SLC7A5 variable showed a degree of accuracy (AUC =0.822; AUC =0.826; AUC =0.757) (Figure 4A,B,I), but had a lower accuracy for HER2 status (AUC =0.523) (Figure 4C). In differentiating PAM50 subtypes, such as the presence of luminal A vs. basal-like subtypes, the predictive ability of the SLC7A5 variables had a high accuracy (AUC =0.929) (Figure 4D), while it had a good degree of accuracy for distinguishing the diverse subtypes of BC: luminal A vs. HER2 (AUC =0.806) (Figure 4E), HER2 vs. basal-like (AUC =0.783) (Figure 4H), luminal B vs. basal-like (AUC =0.781) (Figure 4G), and luminal A vs. luminal B (AUC =0.730) (Figure 4F).
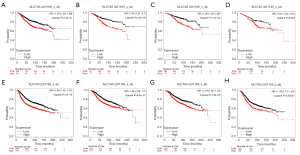
The prognostic value of SLC7A5 in BC patients
For the purpose of determining the prognostic relevance of SLC7A5 in BC, Kaplan-Meier curves were created to evaluate the clinical pattern of SLC7A5 and its expression levels on survival (OS and RFS) and clinical follow-up period. Log-rank tests were conducted for evaluation of statistical significance. The results suggested that higher levels of SLC7A5 were correlated with better OS (n=61, P<0.001; Figure 5), suggesting that the survival times in patients with a higher expression of SLC7A5 were significantly shorter than those patients with lower levels of SLC7A5 expression. We then analyzed SLC7A5 expression as a significant prognostic parameter in the OS (HR =1.64, 95% CI, 1.36–1.99, P<0.001) and RFS value (HR =1.58, 95% CI, 1.42–1.75, P<0.001) of BRCA (Figure 5A,E). In the ER-positive subgroup (OS value: HR =1.95, 95% CI, 1.41–2.7, P<0.001), (RFS value: HR =1.46, 95% CI, 1.25–1.7, P<0.001) (Figures 5B,F), luminal A subtype (OS value: HR =1.92, 95% CI, 1.38–2.66, P<0.001), (RFS value, HR =1.49, 95% CI, 1.26 –1.76, P<0.001) (Figure 5C,G) and luminal B subtype (OS value: HR =1.54, 95% CI, 1.04–2.27, P<0.001), (RFS value, HR =1.38, 95% CI, 1.15–1.65, P<0.001) (Figure 5D,H).
To examine the risk factors correlated to OS in patients with BC in detail, uni- and multivariate Cox regression models were conducted to determine if a high level of SLC7A5 expression is prognostic for worse OS. Univariate analyses showed that age, PAM50 subtype, T stage, N stage, radiation therapy, and expression of SLC7A5 were prognostic factors for OS (P<0.05). Multivariate analyses showed that SLC7A5 expression was an independent risk factor for tumor prognosis (P=0.006), together with age of patients, T stage, N stage, and radiation therapy (P<0.05). These data suggest that a higher level of SLC7A5 expression was associated with decreased OS in BC patients (Table 2).
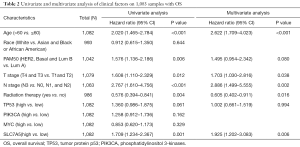
Full table
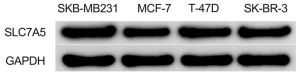
SLC7A5 is upregulated in BC cell lines
We examined the expression of SLC7A5 in BC cell lines, which indicated that the mRNA and protein expression of SLC7A5 were markedly increased in the MDA-MB-231, MCF-7, and T47-D cell lines (Figure 6).
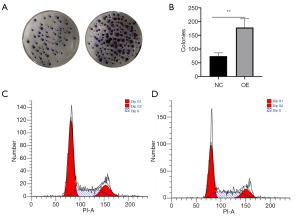
SLC7A5 promoted BC cell proliferation
We then investigated the functional roles of SLC7A5 in BC. SLC7A5 lentiviruses were introduced into MCF-7 cells and increased expression was detected. The colony formation assay revealed that overexpression of SLC7A5 led to an increased proliferative ability of MCF-7 cells (Figure 7A,B). To further explore the impact of SLC7A5 in luminal cell cycle in the control group, we suppressed the expression of SLC7A5 in MCF-7 cells, and conducted cell cycle evaluation by flow cytometry post transfection. Overexpression of SLC7A5 led to a lower number of transfected cells in the G1 phase, and a higher number in the S phase in comparison with the control group (Figure 7C,D). These results further suggested that SLC7A5 is implicated in the tumor growth and proliferation of BC cells.
SLC7A5 activated AKT/mTORC1 pathway by phosphorylation
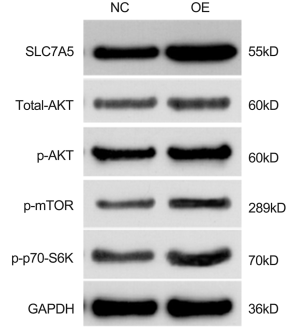
AKT/mTORC1 signaling pathway is known to have a crucial role in multiple tumor types, and suppression of AKT/mTORC1 signaling may limit cancer cell proliferation. To elucidate the mechanism of SLC7A5 involved in regulating cell cycle, we examined relevant proteins through western blotting. We transfected cells with SLC7A5-overexpressing lentiviruses, and detected the protein expression of p-AKT and p-mTOR (Figure 8). As expected, overexpression of SLC7A5 increased the levels of p-AKT and p-mTOR but not the total AKT. The overexpression of SLC7A5 also led to increased p70-S6K mRNA and the phosphorylation of the protein in MCF-7 cells. These results further indicated that SLC7A5 has a promoting function in the growth of luminal BC cells.
Discussion
BC is a highly heterogeneous disease consisting of various different tumor subtypes (9,17) and variations in terms of cellular, molecular, and biological profiles in its response to treatment effects and clinical behaviors. Moreover, various subtypes exhibit disparate nutritional and metabolic needs. The ER+/luminal subtype is the most common subtype of BC (18), and tumors in the subgroup are complicated with regard to diagnosis, prognosis, recurrence rate, and mortality (19). Therefore, exploring the biology and the metabolic pathways of BC subclasses could help to identify potential precision therapeutic targets.
Reprogramming of metabolism is a cancer characteristic in which tumor cells alter the metabolism for the purpose of meeting the needs of cellular building blocks and acquiring the necessary energy (20). Oncogenic activities force the dysregulation of metabolic pathways to build cells increased survival and proliferation in the cancer microenvironment (8). As is well known, AA transport systems are very important to maintaining the proliferation of tumor cells, as they supply the AAs needed for protein synthesis and activate nutritional signaling through the mTORC1 pathway. Our research suggests that the AA transporter, SLC7A5, is important in BC, especially in ER-positive tumors.
Our study showed that in genetic variation, deletion of SLC7A5, rather than its amplification, was significantly correlated to invasive lobular histology. In addition, SLC7A5 expression level in lobular carcinoma was when compared to that in invasive ductal carcinoma, confirming that CNAs of SLC7A5 are present in various BC phenotypes.
SLC7A5 is highly expressed in multiple tumor tissues and in different cell lines (21). The present analysis used TCGA populations to identify any significant associations between the expression level of SLC7A5 and clinical outcomes and pathological parameters, including PAM50 subtype, T stage, N stage, and radiation therapy. The ability of SLC7A5 in predicting the outcome of BC subclasses was found to be highly similar to that of Ki-67. This is in accordance with previous publications which suggested that these 2 parameters are significantly correlated to prognosis in patients with tongue cancer (11), neuroendocrine carcinoma of the lung (22), and thymic carcinoma (23), showing that SLC7A5 is of vital importance in cancer cell proliferation.
SLC7A5 expression was remarkably decreased in low proliferation ER-positive tumors (luminal A subtype) than in luminal B tumors. SLC7A5 expression was more correlated with worse clinical outcomes, such as shorter OS or RFS, in the ER-positive subgroup than in the ER-negative subgroup. This finding may be explained by different energy and nutritional demands for cell growth, proliferation, and tumor microenvironment (TME). Bartlett et al. (24) suggested that SLC7A5 be one part of the 5-gene Mammostrat® immunohistochemistry panel, as it has an increased level of expression. SLC7A5 was correlated with OS, RFS, and distant metastasis-free survival (DMFS) in ER-positive BC patients undergoing endocrine treatment (25). SLC7A5 mRNA and protein expression is much higher in the triple-negative BC and HER2-positive tumors but is not correlated with survival (26). The level of SLC7A5 protein expression has been used as an prognostic factor in ER-positive low-proliferation tumors in our results. We also found the larger sample size of patients with ER-positive tumors has a better potential to reflect a poor prognosis than does a smaller sample size of ER-negative tumors.
An increasing amount of evidence shows that the activation of AKT/mTORC1 pathway is crucial to the cell proliferation process and prevents apoptosis in tumor cells (26). The vital function of SLC7A5 includes supplying essential AAs for protein synthesis and for regulation of mTORC1 signaling (e.g., leucine). The activation mTORC1 in turn regulates protein translation, cell survival and cell proliferation, and prevents apoptosis in malignancies (27,28). Mechanistically, phosphorylation of AKT and mTORC1 is required to activate the pathway to control motility and phenotype transformation of cells. One study showed that the high expression of SLC7A5 activated AKT/mTOR signaling pathway in NSCLC (29). In our analysis, SLC7A5 overexpression significantly elevated the levels of phospho-AKT, phospho-mTOR, phospho-p70-S6K. Up-regulated SLC7A5 through AKT/mTORC1 signaling pathway, which promoted proliferation of MCF-7 to S phase.
In conclusion, our results suggest that the level of the AA transporter SLC7A5 expression is increased in breast cancer tissues and cell lines with differential expression in diverse subtypes. We also found that the level of SLC7A5 expression was correlated with poor prognostic characteristics and worse survival in the ER+/luminal subtypes. Furthermore, overexpression of SLC7A5 was revealed to be important in the proliferation and cell cycle regulation of the MCF-7 cells through its action of upregulating the expression of phospho-AKT, phospho-mTOR, and phospho-p70-S6K. SLC7A5 was thus promoting cell proliferation via activation of AKT/mTORC1 pathway through phosphorylation Therefore, SLC7A5 is a potential prognostic marker and a potential therapeutic target of the luminal subtype of BC treatment.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 81372152, 81800197).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-2247
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-2247
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-2247). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Women’s Hospital of Nanjing Medical University (No.: KY2021054) and informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Ciriello G, Gatza ML, Beck AH, et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015;163:506-19. [Crossref] [PubMed]
- Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009;27:1160-7. [Crossref] [PubMed]
- Jewell JL, Kim YC, Russell RC, et al. Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science 2015;347:194-8. [Crossref] [PubMed]
- Kanai Y, Segawa H, Miyamoto K, et al. Expression cloning and characterization of a transporter for large neutral amino acids activated by the heavy chain of 4F2 antigen (CD98). J Biol Chem 1998;273:23629-32. [Crossref] [PubMed]
- Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol 2014;24:400-6. [Crossref] [PubMed]
- Kimball SR. Regulation of translation initiation by amino acids in eukaryotic cells. Prog Mol Subcell Biol 2001;26:155-84. [Crossref] [PubMed]
- Yanagida O, Kanai Y, Chairoungdua A, et al. Human L-type amino acid transporter 1 (LAT1): characterization of function and expression in tumor cell lines. Biochim Biophys Acta 2001;1514:291-302. [Crossref] [PubMed]
- Abd El-Rehim DM, Ball G, Pinder SE, et al. High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int J Cancer 2005;116:340-50. [Crossref] [PubMed]
- Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346-52. [Crossref] [PubMed]
- Kaira K, Oriuchi N, Imai H, et al. L-type amino acid transporter 1 (LAT1) is frequently expressed in thymic carcinomas but is absent in thymomas. J Surg Oncol 2009;99:433-8. [Crossref] [PubMed]
- Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1-6. [Crossref] [PubMed]
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509-14 [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Gyorffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010;123:725-31. [Crossref] [PubMed]
- Dawson SJ, Rueda OM, Aparicio S, et al. A new genome-driven integrated classification of breast cancer and its implications. EMBO J 2013;32:617-28. [Crossref] [PubMed]
- Rakha EA, El-Sayed ME, Green AR, et al. Biologic and clinical characteristics of breast cancer with single hormone receptor positive phenotype. J Clin Oncol 2007;25:4772-8. [Crossref] [PubMed]
- Senkus E, Kyriakides S, Ohno S, et al. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2015;26:v8-30. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Napolitano L, Galluccio M, Scalise M, et al. Novel insights into the transport mechanism of the human amino acid transporter LAT1 (SLC7A5). Probing critical residues for substrate translocation. Biochim Biophys Acta Gen Subj 2017;1861:727-36. [Crossref] [PubMed]
- Toyoda M, Kaira K, Ohshima Y, et al. Prognostic significance of amino-acid transporter expression (LAT1, ASCT2, and xCT) in surgically resected tongue cancer. Br J Cancer 2014;110:2506-13. [Crossref] [PubMed]
- Wang Q, Holst J. L-type amino acid transport and cancer: targeting the mTORC1 pathway to inhibit neoplasia. Am J Cancer Res 2015;5:1281-94. [PubMed]
- Bartlett JM, Thomas J, Ross DT, et al. Mammostrat as a tool to stratify breast cancer patients at risk of recurrence during endocrine therapy. Breast Cancer Res 2010;12:R47. [Crossref] [PubMed]
- Scope A, Essat M, Pandor A, et al. Gene Expression Profiling and Expanded Immunohistochemistry Tests to Guide Selection of Chemotherapy Regimens in Breast Cancer Management: A Systematic Review. Int J Technol Assess Health Care 2017;33:32-45. [Crossref] [PubMed]
- El Ansari R, Craze ML, Miligy I, et al. The amino acid transporter SLC7A5 confers a poor prognosis in the highly proliferative breast cancer subtypes and is a key therapeutic target in luminal B tumours. Breast Cancer Res 2018;20:21. [Crossref] [PubMed]
- Liang Z, Cho HT, Williams L, et al. Potential Biomarker of L-type Amino Acid Transporter 1 in Breast Cancer Progression. Nucl Med Mol Imaging 2011;45:93-102. [Crossref] [PubMed]
- Singh N, Scalise M, Galluccio M, et al. Discovery of Potent Inhibitors for the Large Neutral Amino Acid Transporter 1 (LAT1) by Structure-Based Methods. Int J Mol Sci 2018;20:27. [Crossref] [PubMed]
- Cheng SW, Fryer LG, Carling D, et al. Thr2446 is a novel mammalian target of rapamycin (mTOR) phosphorylation site regulated by nutrient status. J Biol Chem 2004;279:15719-22. [Crossref] [PubMed]
(English Language Editor: J. Gray)


