
Six ALPL gene variants in five children with hypophosphatasia
Introduction
Hypophosphatasia (HPP) (MIM 146300, 241500, 241510) is a rare hereditary disease that is characterized by defective mineralization of bones and teeth and decreased serum tissue nonspecific alkaline phosphatase (TNSALP) activity. It was first described by Chown in 1936 and named by John Campbell Rathbun, a Canadian physician, as hypophosphatasia in 1948. HPP has diverse clinical manifestations. Mild cases may present only with premature tooth loss, reduced dental bone mass, and enlarged pulp cavities but no skeletal abnormalities, while in severe cases there may be severe skeletal abnormalities, vitamin B6–dependent seizures, secondary severe infections, respiratory failure, and hypercalcemia (which can lead to death). According to genetic variants found in specific populations, the incidence of HPP varies greatly, but it is estimated that the incidence of live births ranges from 1/2,500 to 1/300,000 (1,2).
HPP is a genetic condition associated with mutations in the liver/bone/kidney alkaline phosphatase gene (ALPL gene) which encodes the tissue-nonspecific alkaline phosphatase (TNSALP). It can be inherited in an autosomal dominant or an autosomal recessive manner. Located on chromosome 1p36.1-p34, ALPL gene is composed of 12 exons and encodes TNSALP, a phosphomonoesterase that consists of 524 amino acids and that contains active site, active vally, homodimer interface, crown domain, and calcium-binding site (3). As a homodimer, ALPL is attached to the cellular membrane through a glycophosphatidylinositol (GPI) anchor at the C-terminus, orienting the active sites toward the extracellular environment to exert their effects (4). ALPL is widely present in various tissues, especially in bones, liver, kidneys, and developing teeth. TNSALP generates Pi by hydrolyzing PPi (inorganic pyrophosphate), which participates in the formation of hydroxyapatite crystals, and promotes the mineralization of osteoblasts. In addition, TNSALP forms pyridoxal (Pyridoxal, PL) by hydrolyzing PLP, which acts as a cofactor in the enzyme reaction and participates in the metabolism of transaminase and certain neurotransmitters. Any ALPL variant that can affect the expression, folding, modification, and trafficking of the TNSALP protein, resulting in HPP (5). Here we performed high-throughput sequencing analysis in 5 children with clinically suspected HPP and identified 6 variants in the ALPL gene. Also, we investigated their pathogenic mechanisms.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-21-2096).
Methods
Clinical data
Five children (three males and two females, with ages ranging from 6 days to 2 years and 5 months) with clinically diagnosed HPP were enrolled. Three patients had infantile HPP, with defective bone mineralization (e.g., skeletal deformities) and convulsions as the main clinical manifestations; two children had childhood HPP, with short stature and rickets-like X-ray findings as the main clinical symptoms. The clinical data and genetic variants of these 5 HPP patients have been described in a previous article (Table 1) (6).
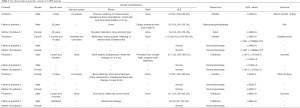
Full table
Ethical statement and genetic testing
The study was approved by the ethical board of our center (ethical approval No. 2019; NLS research No. 232), and informed written consent was obtained from the parents of each of the 5 children. This study conformed to the provisions of the Declaration of Helsinki (as revised in 2013).
The peripheral blood samples were collected from the children and their parents and sent to a third-party testing company for whole-exome sequencing. The TruSeq Exome Library Prep Kit (Illumina) was used for capturing the whole genome and constructing the sequencing library. Sequencing was performed on the HiSeq 2500 sequencing platform by using the HiSeq SBS Kit v4 kits (Illumina). The sequencing data were subjected to bioinformatic analysis, pathogenicity prediction (with SIFT, Polyphen2, MutationTaster, and PROVEAN, etc.), clinical symptom comparison/pedigree analysis, and database search, and literature review for relevant diseases. The candidate gene variants were classified according to American College of Medical Genetics (ACMG) and American Society of Molecular Pathology (AMP) guidelines and validated by chain termination method (Sanger method). The effects of the screened variants on the proteins were predicted by using the PROVEAN (Protein Variation Effect Analyzer) software.
Plasmid construction
The generation of an expression vector including the complete coding region of the human tissue nonspecific alkaline phosphatase (ALPL; GenBank accession no. NM_000478), designated TNAP wild type (7), has been described in detail by Mentrup et al. Accordingly, we constructed the plasmids for the ALPL gene variants by using the GeneCopoeia technology.
Transfection and enzyme activity assay
HEK293 cells were transfected with the constructed plasmids expressing wild-type (WT) and mutant ALPL in Lipofectamine 2000 (Thermo Fisher Scientific). Cells were collected at 24, 48, and 72 hours to extract total cellular protein. The enzyme activity of TNSALP was measured by using an alkaline phosphatase assay kit (Beyotime). The experiments were repeated 3 or more times.
Statistical analysis
The TNSALP enzyme activity of the same transfected plasmid at different culture time and the TNSALP enzyme activity of different plasmids transfected at the same culture time were analyzed statistically. If P≤0.05, the difference was statistically significant.
Immunofluorescence test
HEK293 cells were transfected with plasmids expressing wild-type and mutant ALPL in Lipofectamine 2000. Twenty-four hours after transfection, the cells were washed 3 times with phosphate-buffered saline (PBS) and fixed with ice-cold methanol for 10 minutes. After the removal of methanol, the cells were blocked with 3% bovine serum albumin (BSA; prepared with PBS) for 30 minutes at room temperature and washed 3 times with PBS. The primary antibody (1:50, AP1474C, Abgent Inc.) was incubated at 4 °C for 16 hours and washed 3 times with PBS. The secondary antibody [1:200, fluorescein isothiocyanate (FITC) antimouse immunoglobin G; Proteintech] was incubated at room temperature for 2 hours and washed 3 times with PBS. DAPI (Solarbio) was used to stain cell nuclei at room temperature for about 10 minutes, washed 3 times with PBS, and then stored in PBS at 4 °C. The blue (DAPI) fluorescence and the green (FITC) fluorescence were observed under an inverted fluorescence microscope (UltraVIEW VoX & IX81, Perkin Elmer & Olympus), where the green fluorescence showed the target protein TNSALP and the blue fluorescence showed the nuclei.
Western blotting
Cells were collected and the total proteins were extracted and loaded (20–40 µL for each group of proteins). Bicinchoninic acid (BCA) assay was performed before loading. After 10% polyacrylamide gel electrophoresis (SDS-PAGE), transfer to polyvinylidene fluoride (PVDF) membrane, and blockage with 5% defatted milk for 2 hours, the samples were inoculated overnight at 4 °C with the specific antibody (1:2,000; ALPL Antibody, AP1474C, Abgent). The next day, after washing with 0.1% tris-buffered saline with Tween20 (TBST) 3 times, the samples were mixed with secondary antibody (1:2,000; goat antirabbit, ProteinTech) and incubated at 37 °C for 1 hour. Subsequently, the samples were washed with TBST 3 times and developed with enhanced chemiluminescence (ECL). The band density was quantified using Fusion software (Vilber). The experiments were repeated 3 or more times.
Results
Six ALPL variants identified
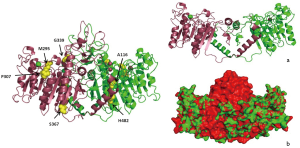
A total of 6 ALPL variants were identified in 5 HPP children (Table 2). Sanger sequencing showed heterozygous autosomal dominant variants in proband 1, with the mother being an HPP patient, and the variants having come from the mother. Compound heterozygous variants were identified in probands 2 and 3, in whom the disease was autosomal recessive, and whose parents were all carriers. A homozygous variant was identified in proband 4, in whom the disease was autosomal recessive, with both parents being carriers. Heterozygous autosomal dominant variants were found in proband 5, whose father was an HPP patient, with the variants having come from the father. No de novo mutation was found. Among these 6 variants, c.1014_c.1015ins G, c.1097_c.1099del CCT, and c.1446C>A were not found in the 1000 Genomes Project, dbSNP (The Single Nucleotide Polymorphism Database), or the HapMap project, and therefore are reported here for the first time. They were predicted to be deleterious by in silico analysis and were potentially pathogenic according to the American College of Medical Genetics and Genomics (ACMG) criteria. The 3D structure of the protein was predicted by homology modeling using crystal structure of AP from Human Placenta (8) as a template (Figure 1).

Full table
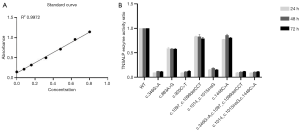
Differential effects of different variants on the alkaline phosphatase activity of their expressed proteins
The wild-type ALPL, c.346G>A, c.883A>G, c.920C>T, c.1097_c.1099del CCT, c.1014_c.1015ins G, c.1446C>A, c.346G>A+c.1097_c.1099delCCT, and c.1014_ c.1015ins G+c.1446C>A were overexpressed in the cells, and TNSALPase activity was detected 24, 48, and 72 hours after the HEK-293T cells were transfected. The standard curves of the standard products were obtained (Figure 2A). The sample enzyme activity was normalized by the sample protein concentration, and the results of enzyme activity determination are shown in Figure 2B. The enzyme activity level in each group reached the highest level 24 hours after transfection, and the differences in enzyme activity among the 3 time points within each group were not statistically significant. The wild type had the highest enzyme activity, and the TNSALP enzyme activities in c.346G>A, c.920C>T, c.1014_c.1015ins G, c.346G>A + c.1097_c.1099del CCT, and c.1014_c.1015ins G + c.1446C>A groups decreased to 9.22%, 8.86%, 15.43%, 9.08%, and 8.91% of the wild type, respectively. In addition, the TNSALPase activities in the c.883A>G, c.1097_c.1099del CCT, and c.1446C>A groups decreased to 58.65%, 83.23%, and 77.60% of the wild type, respectively. Thus, different variants had different effects on TNSALPase activity, among which c.346G>A, c.920C>T, c.1014_c.1015ins G, c.346G>A + c.1097_c.1099del CCT, and c.1014_c.1015ins G + c.1446C>A had remarkable effects on enzyme activity, and more than 80% of enzyme activity was lost. The c.883A>G, c.1097_c.1099del CCT, and c.1446C>A variants had smaller effects on the enzyme activity, and the majority of the enzyme activity was retained. The effects of compound heterozygous variants on enzyme activity were consistent with the effect of the compound heterozygous variant having a greater effect on enzyme activity.
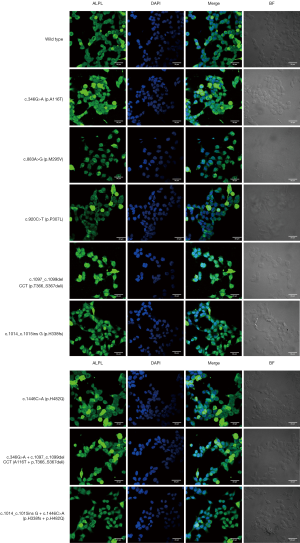
Variants caused a decrease in alkaline phosphatase expression in the cell membrane
Blue (DAPI) fluorescence and the green (FITC) fluorescence were observed under an inverted fluorescence microscope, where the green fluorescence showed the target protein TNSALP and the blue fluorescence showed the nuclei. As shown in Figure 3, different variants (especially c.883A>G and c.920C>T) caused different degrees of decrease in alkaline phosphatase expression in the cell membrane.
The protein expressions of variants
The protein expressions of all variants were lower than the expression of the wild type (Figure 4), but the degrees of expression reduction varied across the different variants. Except for 1014-G-1015 + C1446A, the clinical phenotypes the variants were negatively correlated with the protein expression levels.
Plasmids expressing ALPL wild type, and each variant were transfected into HEK-293T cells, and the proteins were collected after 24 hours for Western blotting. In lanes corresponding to the wild type, and A883G, △1097–1099, and C1446A variants, significant protein bands were found at 80 and 66 kDa, with the wild type having the strongest bands. In these 5 children and their families, A883G showed autosomal dominant inheritance, and the patients had mild rickets-like clinical manifestations; △1097–1099 and C1446A showed autosomal recessive inheritance, and their carriers had a normal phenotype. G346A also showed two bands, but the band at 80 kDa was significantly attenuated; its carriers had mild or severe clinical manifestations, and the mode of inheritance was autosomal dominant. C920T only had a markedly attenuated band at 80 kDa, and its carriers had a normal clinical presentation, with a severe clinical phenotype in homozygous patients. For 1014-G-1015 + C1446A, no protein band was seen at 80 and 66 kDa, but the patient only had dental defects and rickets-like changes. For 1014-G-1015 and △1097–1099 + G346A, only an attenuated band was observed at 66 kDa; the patient with 1097-1099 + G346A had severe clinical manifestations, whereas the 1014-G-1015 carrier had a normal clinical phenotype. Therefore, the protein expression levels were decreased in the variant groups rather than in the wild-type groups. However, the reduction in expression differed among the variant types. Most of the clinical phenotypes were negatively correlated with the protein expression levels. Notably, however, although some variants lead to significantly reduced protein expression, the clinical phenotypes were mild; thus, different mutations may lead to the same protein expression changes, but the clinical manifestations may vary widely.
Discussion
In the present study, 3 novel variants were identified in 5 HPP children, complementing the human ALPL gene mutation database. TNSALP enzyme activity was differentially down regulated by different ALPL mutations. The genotype and phenotype of HPP are highly variable, and the mild and severe phenotypes of the same variant also vary among different patients.
HPP is a genetic disease caused by defects in bone tissue mineralization and classified into at least 6 forms according to the age of onset and severity: perinatal (lethal), perinatal (benign), infantile, childhood, adult, and odontohypophosphatasia. The prognosis of HPP is related to the time of onset. The earlier the onset, the worse the prognosis. In a study of 358 patients (Europe, Canada, USA, Australia), the most common forms were perinatal (30.4%) and infantile (23.5%), followed by adult and childhood odontohypophosphatasia (9). According to the data, the statistics of children with HPP in China are mostly case reports and the most common phenotypes of hypophosphatemia in Chinese children were the infant type (43%), followed by the childhood type (23%) (10-22). Severe HPP is mostly inherited in autosomal recessive form, while perinatal (benign), childhood, adult, and odontohypophosphatasia with mild symptoms are usually inherited in autosomal dominant or recessive form. Currently, HPP can be clinically diagnosed based on serum alkaline phosphatase (ALP) activity, urinary phosphoethanolamine (PEA) level, fetal ultrasound, and X-ray, but all of them lack specificity. The ALPL gene test is recommended for high-risk populations with a family history of HPP or for patients with clinically suspected HPP, as it may increase the accuracy of HPP diagnosis. Currently, genetic testing can screen 95% of patients with severe HPP (23) and is a more reliable diagnostic tool than serum ALP testing. According to the data, a total of 55 ALPL gene variants have been reported in China (10-22), 65.4% of which are missense variants and 23.6% of which are deletions and insertions, with the remaining variants consisting of nonsense variants (3.6%), splicing mutations (7.3%), and 50 variants located in exons and 5 in introns. Notably, 6 cases had different variants in the c.407 locus. Whether there are hotspots for the ALPL variants warrants further investigation through large-sample, multicenter studies. The variants found in our current study included missense variants in 4 cases, a small deletion variant in 1 case, and an insertion variant in 1 case.
In our current analysis, the variant in proband 1 was c.346G>A, which caused a change from alanine to threonine at position 116, disrupting the hydrogen bond structure at the homodimer interface. As a result, the protein could not fold correctly and form a disulfide-bonded dimer. Western blotting revealed a significantly weaker band at 80 kDa, and immunofluorescence showed an increased expression in cytoplasm in the variant group, but it did not completely reach the cell membrane. In addition, TNSALPase activity significantly declined. The pathogenicity of variants in this position has been previously reported as odontohypophosphatasia, which is autosomal dominant, and some investigators have also found that the variant in this position confers a dominant negative effect (24). In the present study, the mother of proband 1 had the same genotype as proband 1; however, the mother had reduced ALP levels and presented with early tooth loss, bone pain, and joint dislocation, and the disease was classified as mild adult HPP; in contrast, the child had the clinical manifestations of severe infantile HPP. Thus, the clinical phenotypes were not consistent. The father had dental involvement and reduced serum ALP in peripheral blood, but the ALPL gene testing showed a negative result. Thus, further examinations are required to rule out the possibility that the negative result was due to variants in the nonnucleotide base sequences and/or the blind area of intron regulatory regions. Similarly, Mentrup et al. have proposed that if variants cannot be detected by conventional coding methods (including coding sequences and exon-intron boundaries), the key regulatory motifs (such as branch point sequences) need to be investigated more thoroughly to confirm a genetic diagnosis (25). Meanwhile, the contributions of environmental factors to individual differences cannot be neglected, and further mechanistic studies are required in this area.
Proband 2 was presented to a local hospital due to irritability, crying, and repeated convulsions, where he was incorrectly diagnosed as with “intracranial hypertension syndrome: atypical bacterial meningitis” and treated accordingly. The wrong diagnosis was made due to the lack of a thorough review of clinical manifestations, laboratory tests, and imaging findings. According to the literature, the accumulation of pyridoxal phosphate (PLP), the active form of vitamin B 6, due to reduced TNSALP levels may lead to seizures in some infants with HPP and in experimental ALPL-knockout mice (26). Therefore, infantile HPP usually has clinical manifestations dominated by neurological symptoms such as convulsions within the first 6 months of life (27,28); furthermore, convulsions are the most important neurological clinical manifestation of heavy HPP (28), with a mortality rate of 50% to 90%. Three cases of infantile HPP in our present study had neurological manifestations (e.g., convulsions), and the case-fatality rate reached 100%. Therefore, for children with convulsions, especially those with unexplained recurrent convulsions with skeletal abnormalities, clinicians should be alert to the possibility of HPP and search for the best available evidence. At the same time, hypophosphatemia should be differentiated from diseases such as low-phosphorus anti-D rickets, vitamin D-dependent rickets, achondroplasia and osteogenesis imperfecta. It is very difficult to distinguish them only by phenotype, but they can be distinguished by electrolytes, serum alkaline phosphatase levels and X-ray examination.
The variant c.1097_c.1099del CCT carried by proband 2 and the variants c.1014_c.1015ins G and c.1446C>A carried by proband 3 were among the compound heterozygous variants, which were previously unreported de novo variants. In our experiments, the overexpression of c.1097_c.1099del CCT and c.1446C>A genetic variants decreased the enzyme activities to 83.23% and 77.60% of that of the wild type, respectively. Western blotting revealed obvious protein bands at 80 and 66 kDa, and immunofluorescence assay showed protein expressions on the cellular membrane, which were consistent with the clinical phenotypes of their carriers. Prediction of the 3D structure of c.1014_c.1015ins G protein suggested that this variant could lead to the deletion of the coronal structure domain. In cellular assays, overexpression of this variant could lead to a significant decrease in enzyme activity (15.43%). In Western blotting, there was only an attenuated band at 66 kDa. However, the clinical phenotype of the carrier was normal. A possible explanation is that this variant disrupts posttranslational folding and forms defective protein monomers, while the mutant protein monomers fail to assemble into heterodimers that interfere with the wild-type protein products. It is speculated that this variant has no dominant negative effect, but this of course, needs to be further validated.
Severe forms of HPP are usually inherited as autosomal recessive traits with compound heterozygosity for 2 missense mutations. The residual enzyme activity after missense mutations depends on the alterations of the mutated amino acids and the effects of the mutant amino acid residues, and sometimes there is a dominant negative effect caused by mutations. It has been reported that certain variants do not produce dominant negative proteins in heterozygous carriers (29), but some specific conditions (e.g., lactation) seem to trigger clinical symptoms. In contrast, for autosomal dominant variants, the severity of clinical manifestations varies among individuals carrying the same single heterozygous variant. In addition to alterations in noncoding sequences and the blind area of intron regulatory region sequencing, it is also important to be alert to incomplete penetrance and variable expressivity due to environmental and individual factors. Currently, it is difficult to reliably predict the severity of phenotypes based on genotype and mode of inheritance. Environmental factors, epigenetics, and regulatory genes should also be considered when analyzing the impact of de novo variants (30). Studies have shown that some variants of the ALPL gene exhibit a dominant negative effect, although the specific hotspots where dominant negative effects may occur remain unclear. The chance for the same variant to exert its dominant negative effect varies across individuals, and the specific mechanisms need to be further investigated.
The variants in probands 4 and 5 were c.920C>T and c.883A>G, respectively, and the amino acid alterations were p.P307L and p.M295V, respectively. Both variants were located in the calcium-binding region. There are 5 amino acid residues (Trp270, Arg272, Leu275, Asp306, and Glu311) in the calcium-binding region that have been highly conserved throughout mammalian evolution, strongly suggesting that they have important roles in enzyme structure and function (31). Proband 4 had a variant of c.920C>T, with amino acid alteration site p.P307L adjacent to the highly conserved site Asp306 in this region. It is assumed this variant has greater effects on TNSALP structure and function. In the present study, c.920C>T overexpression resulted in a significant reduction in enzyme activity (8.86%); in western blotting, there was an attenuated protein band at 80 kDa and a barely visible band at 66 kDa; also, immunofluorescence showed a significant reduction in protein expression on cell membranes. Born to inbreeding, the proband 4 had severe infantile HPP. Both of his parents were heterozygous carriers of the variant, but did not show any clinical manifestations. In a boy with mutation (c.920C>T) in the ALPL gene, the clinical manifestations included bulging anterior fontanelle, growth failure, nephrocalcinosis, and impaired bone mineralization (32), which is consistent with our findings. In contrast, for the variant c.883A>G that is also located at the calcium-binding site, the amino acid alteration site p.M295V is distant from the conserved site. Overexpression of this variant led to only mildly impaired enzyme activity, and the clinical phenotype was mild childhood HPP. Therefore, when analyzing the effects of de novo variants in ALPL gene, attention should be paid to the relationships between amino acid alterations caused by a specific variant with the key structural domains and the highly conserved regions.
In our current study, 3 novel variants were identified in 5 HPP children that were studied, and their discovery can hopefully enrich the human ALPL gene mutation database. Different mutations in the ALPL gene can downregulate the activity of TNSALP enzyme (and thus affect its function) by affecting protein expression and translational modifications. The same variant may cause clinical manifestations of varying severities in different individuals due to the presence of dominant negative effects, alterations in noncoding sequences, blind area of intron regulatory region sequencing, and variations in environmental and individual factors. At present, the statistics of children with HPP in China are mostly case reports, and there is a lack of multi-center large-scale research data. The clinical, genetic characteristics and follow-up data of HPP patients in China need to be further summarized and studied. At the same time, standardized management of children with HPP should be carried out, and regular follow-up and evaluation should be strengthened. For families with children with HPP, routine prenatal diagnosis should be recommended. These are what we need to do in the next step.
Acknowledgments
We greatly appreciate our patients and their families for allowing us to use their clinical data and blood samples. The authors also appreciate the academic support from AME Hypophosphatasia Collaborative Group.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-2096
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-2096
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-2096). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was proved by the ethics committee of the Children’s Hospital Affiliated to Chongqing Medical University (ethical approval No. 2019; NLS research No. 232). Informed written consent was obtained from the parents of each of the 5 children. The study conformed to the provisions of the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hypophosphatasia Mornet E. Orphanet J Rare Dis 2007;2:40. [Crossref] [PubMed]
- Mornet E, Yvard A, Taillandier A, et al. A molecular-based estimation of the prevalence of hypophosphatasia in the European population. Ann Hum Genet 2011;75:439-45. [Crossref] [PubMed]
- Whyte MP. Physiological role of alkaline phosphatase explored in hypophosphatasia. Ann N Y Acad Sci 2010;1192:190-200. [Crossref] [PubMed]
- Hollis A, Arundel P, High A, et al. Current concepts in hypophosphatasia: case report and literature review. Int J Paediatr Dent 2013;23:153-9. [Crossref] [PubMed]
- Sultana S, Al-Shawafi HA, Makita S, et al. An asparagine at position 417 of tissue-nonspecific alkaline phosphatase is essential for its structure and function as revealed by analysis of the N417S mutation associated with severe hypophosphatasia. Mol Genet Metab 2013;109:282-8. [Crossref] [PubMed]
- Su N, Zhu M, Xu K, et al. Clinical and genetic analysis of hypophosphatasia in 5 children. Journal of Clinical Pediatrics 2020;38:289-93.
- Mentrup B, Marschall C, Barvencik F, et al. Functional characterization of a novel mutation localized in the start codon of the tissue-nonspecific alkaline phosphatase gene. Bone 2011;48:1401-8. [Crossref] [PubMed]
- Le Du MH, Stigbrand T, Taussig MJ, et al. Crystal Structure of Alkaline Phosphatase from Human Placenta at 1. 8 A Resolution: Implication for a substrate specificity. J Biol Chem 2001;276:9158-65. [Crossref] [PubMed]
- Fauvert D, Brun-Heath I, Lia-Baldini AS, et al. Mild forms of hypophosphatasia mostly result from dominant negative effect of severe alleles or from compound heterozygosity for severe and moderate alleles. BMC Med Genet 2009;10:51. [Crossref] [PubMed]
- Li L, Chen B, Yan F. Characteristics of Gene Mutation and Chief Complaint of Chinese Patient with Hypophosphatasia. Journal of Medical Research 2016;4508:74-9.
- Bai Y, Liu N, Yang J, et al. Mutation analysis for two hypophosphatasia families with targeted next-generation sequencing. National Medical Journal of China 2016;96:3718-23. [PubMed]
- Wang Wenjie, Fu Wenzhen, He Jinwei, et al. Adult hypophosphatasia caused by ALPL mutation: one pedigree study. Chin J Endocrinol Metab 2017;33:585-9.
- Li DF, Lan D, Zhong JZ, et al. Infantile hypophosphatasia caused by a novel compound heterozygous mutation: a case report and pedigree analysis. Chinese Journal of Contemporary Pediatrics 2017;19:539-44. [PubMed]
- Zhou W, Li L, Chen B, et al. Hypophosphatasia: a case report and gene mutation analysis. Chinese Journal of Stomatology 2017;52:578-9. [PubMed]
- Lu W, Shi C, Cai D, et al. Clinical and genetic analysis of a family with low alkaline phosphatase. Journal of Clinical Pediatrics 2017;35:682-6.
- Wan J, Zhang L, Liu T, et al. Genetic evaluations of Chinese patients with odontohypophosphatasia resulting from heterozygosity for mutations in the tissue non specific alkaline phosphatase gene. Oncotarget 2017;8:51569-77. [Crossref] [PubMed]
- Huang H. Molecular research of pathogenic mutations for a Hypophosphatasia pedigree Identification of causative gene and functional study for a pedigree with Hypertrophic Cardiomyopathy. Guangzhou: Southern Medical University, 2017.
- Zhang L, Gu Y, Wang S, et al. Analysis of genetic variation in a family with dental hypoalkaline phosphatasemia. Chinese Journal of Practical Internal Medicine 2018;38:385-7.
- Li P. A Family of Hypophosphatasia (HPP). Fuzhou: Fujian Medical University, 2018.
- Dai Q, Yan R, Zhang J. One case of perinatal lethal hypophosphatasia with X-ray manifestations and literature review. Clinical Research and Practice 2019;4:9-10.
- Lu J, Huai L, Lu C, et al. Prenatal diagnosis and genetic analysis of 17 fetuses with skeletal dysplasia. Chinese Journal of Medical Genetics 2020;37:1217-21. [PubMed]
- Yu Y, Chen A, Zheng J, et al. One patient with perinatal hypophosphatasia due to mutations in the tissue-nonspecific alkaline phosphatase gene. Chinese Journal of Endocrinology and Metabolism 2020;36:321-5.
- Watanabe A, Yamamasu S, Shinagawa T, et al. Prenatal genetic diagnosis of severe perinatal (lethal) hypophosphatasia. J Nippon Med Sch 2007;74:65-9. [Crossref] [PubMed]
- Ishida Y, Komaru K, Oda K. Molecular characterization of tissue-nonspecific alkaline phosphatase with an Ala to Thr substitution at position 116 associated with dominantly inherited hypophosphatasia. Biochim Biophys Acta 2011;1812:326-32. [Crossref] [PubMed]
- Mentrup B, Girschick H, Jakob F, et al. A homozygous intronic branch-point deletion in the ALPL gene causes infantile hypophosphatasia. Bone 2017;94:75-83. [Crossref] [PubMed]
- Whyte MP, Landt M, Ryan LM, et al. Alkaline phosphatase: placental and tissue-nonspecific isoenzymes hydrolyze phosphoethanolamine, inorganic pyrophosphate, and pyridoxal 5'-phosphate. Substrate accumulation in carriers of hypophosphatasia corrects during pregnancy. J Clin Invest 1995;95:1440-5. [Crossref] [PubMed]
- Gasque KC, Foster BL, Kuss P, et al. Improvement of the skeletal and dental hypophosphatasia phenotype in Alpl-/- mice by administration of soluble (non-targeted) chimeric alkaline phosphatase. Bone 2015;72:137-47. [Crossref] [PubMed]
- Linglart A, Biosse-Duplan M. Hypophosphatasia. Curr Osteoporos Rep 2016;14:95-105. [Crossref] [PubMed]
- Hofmann C, Liese J, Schwarz T, et al. Compound heterozygosity of two functional null mutations in the ALPL gene associated with deleterious neurological outcome in an infant with hypophosphatasia. Bone 2013;55:150-7. [Crossref] [PubMed]
- Mornet E. Genetics of hypophosphatasia. Arch Pediatr 2017;24:5S51-6.
- Hu JC, Plaetke R, Mornet E, et al. Characterization of a family with dominant hypophosphatasia. Eur J Oral Sci 2000;108:189-94. [Crossref] [PubMed]
- Mohn A, De Leonibus C, de Giorgis T, et al. Hypophosphatasia in a child with widened anterior fontanelle: lessons learned from late diagnosis and incorrect treatment. Acta Paediatr 2011;100:e43-6. [Crossref] [PubMed]
(English Language Editor: J. Gray)



