
The therapeutic effects and mechanisms of Long Chai Fang on chronic hepatitis B
Introduction
Chronic hepatitis B (CHB) is an infectious chronic disease caused by liver invasion and damage by the hepatitis B virus (HBV) that causes disabling fatigue, nausea, bloating, hepatalgia, and splenomegaly (1,2). There are approximately 350 million HBV carriers worldwide 20 million of whom are CHB patients and 93 million of whom are chronic carriers in China (3-5). The long-term infection of HBV can lead to hepatic fibrosis, liver cirrhosis, and hepatocellular carcinoma. Interferon (IFN) and nucleoside analogue are clinical anti-HBV drugs used to treat CHB; however, they can cause drug resistance (6). In recent years, traditional Chinese medicine (TCM) has been widely concerned with anti-HBV activity (7). Long Chai Fang (LCF), which was developed by Professor Jinshi, a doctoral advisor at the Nanjing University of TCM and a famous TCM physician, is an empirical prescription used to treat patients with CHB. However, the molecular mechanism of LCF against CHB remains unknown. This study sought to explore the effects and related mechanisms of the LCF treatment of CHB in a duck model. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-21-1923).
Methods
LCF preparation
LCF comprises 7 herbs (i.e., 15 g of Solanum nigrum, 8 g of Bupleurum, 30 g of Hedyotis diffusa, 12 g of Ligustrum lucidum, 9 g of Jiao Shanzhi, 30 g of sarmentosum, and 5 g of licorice). These herbs were obtained from Jiangsu Provincial Academy of TCM and boiled using a TCM method. The water extract was then concentrated using a rotary evaporator, and long-term preserved at −80 °C. The preliminary research of the research group showed that the main effective components of LCF include polysaccharose, saponins, flavonoids, alkaloids, solasonine, and solamargine. Of these, the content of polysaccharide is the highest, followed by total soap.
The results of a quality control (QC) study showed that the paste-out rate of the LCF was 0.0841 g/mL (19.6%), total ash was 0.0161 g/mL (19.2%) of the total solid output, and the protein content was 7.73 mg/mL. The average content of the active components was as follows: 15.672 mg/mL of polysaccharide (18.6% of the total solid output), 9.274 mg/mL of soap (11.0% of the total solid output), 5.756 mg/mL of flavonoids (6.8% of the total solid output), 3.25 mg/mL of alkaloids (3.86% of the total solid output), 0.00929 mg/mL of solenoids (0.011% of the total solid output), 0.00865 mg/mL of Australian eggplant edge alkali (0.010% of the total solid output).
Screening the active ingredients and potential targets of LCF by network pharmacology
In the TCM systems pharmacology (TCMSP) database, LCF, including Hedyotis diffusa, Bupleurum chinense, Sedum sarmentosum Bunge, Glycyrrhiza uralensis, Fisch, cape jasmine fruit, solanum nigrum L, and Fructus Ligustri Lucidi, were retrieved. The TCM integrated database (TCMID), excluding the TCMSP database, was used for retrieval, and China pharmacopoeia and literature were as potential active ingredients. Absorption, distribution, metabolism, and excretion were then used to evaluate the effects and risks of LCF ingredients in vitro. An oral bioavailability of ≥30% and drug-likeness of ≥0.18 were set as the screening conditions for the active ingredients in the TCMSP database. A search of the PubChem database (http://pubchem.ncbi.nlm.nih.gov/) was conducted to obtain the composition structure, and the Swiss Target Prediction database was then searched. The prediction score was eventually made greater than 0 for the drug targets.
The Drugbank, and Genecards databases were then searched using the term “Chronic hepatitis B” to identify disease targets. TheVenny2.1 online software mapping tool was used to analyze the drug and disease targets, draw a Wayne diagram, and identify the drug-disease targets.
Cytoscape software was performed to construct the network diagram of “LCF—component—disease—target”, and the main active components of the TCM compound were subjected to a topological analysis. The degree values indicated the correlations between the components and the targets. Next, 185 common targets were input into the STRING database with the protein “Homo sapiens” and a minimum threshold 0.4. Cytoscape software was used to draw a protein interaction network diagram. Finally, the protein-protein interaction (PPI) network was imported into Cytoscape, and the molecular complex detection module was used for the gene cluster analysis and core target screening.
A Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis was performed to analyze the key target genes. The pathways with the highest enrichment combined using Cytoscape were analyzed to establish the “disease—composition—targets—access network” by screening for possible pathways associated with CHB and by finding the potential active ingredients of LCF in the treatment of CHB.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Reagents
Lamivudine (LAM) (100 mg per tablet,) was produced by GlaxoSmithKline (GSK). A Digoxin DIG labeling kit was purchased from ROCHE Company (US). Tris base, hydrochloric acid, and glucose were purchased from the Shanghai Jerry Limited Liability Company. Lysozyme, maleic acid and Tween-20 were purchased from the US Aladdin Reagent Company. Sodium chloride, magnesium chloride, sodium hydroxide, and sodium acetate were purchased from the Sinopharm Group Company. Sodium lauryl sulfate, glacial acetic acid, and boric acid were purchased from the Nanjing Chemical Reagent Company.
Animal studies
Pekin-Aylesbury cross-bred ducklings, weighing 150±50 g, were purchased from Xinhe Deese and Ducks Farm in Longmiao Town, Shuyang City, Jiangsu Province. They were housed at room temperature (22–25 °C) with a humidity of 40–70%, had free access to water and food, and were placed on a 12 h day and 12 h night cycle. Sixty Duck Hepatitis B virus (DHBV) negative ducklings were inoculated with 30 µL of DHBV-deoxyribonucleic acid (DNA)-positive serum from Shanghai ducks; ducklings with congenital infections of DHBV were excluded. After 7 days of infection (D0), serum samples were collected from the toe vein to detect DHBV, and the DHBV-positive ducklings were divided into the following 5 groups: (I) a model group; (II) a positive control-LAM group (0.2 g/kg); (III) a low-dose LCF (2 g/kg) group; (IV) a medium-dose LCF (15 g/kg) group; and (V) and a high-dose LCF (45 g/kg) group. Intragastric administration once a day for 10 days. The experiment was terminated after 3 days of drug discontinuance. Serum was collected to detect the content of DHBV-DNA by dot-blot hybridization on day 5 (D5), day 10 (D10) and day 3 after the cessation of treatment (P3). Experiments were performed under a project license (No.: AEWC-20181205-65) granted by ethics committee of Jiangsu Province Hospital on Integration of Chinese and Western Medicine, in compliance with Jiangsu Province Hospital on Integration of Chinese and Western Medicine guidelines for the care and use of animals.
Viruses and plasmids
DHBV was extracted from the fresh serum of a 12-month DHBV-positive Shanghai Sheldrake. The DHBV-PUC19 recombinant plasmid and the DHBV probe were provided by the College of Pharmacy, Fudan University.
Dot-blot hybridization
Membrane preparation
The nylon membranes were soaked in 2× SSC (l× of SSC comprises 0.15 M of sodium chloride and 0.015 M of sodium citrate, pH 7.0) and spotted in a 96-well suction filtration device. Fifty µL serum specimens were added to each well, and the membranes were denatured in 1 mol/L NaOH for 30 min at room temperature. The membranes were then neutralized for 15 min with a scrubbing solution [1 M Tris hydrochloride (pH 7.0)] and then immersed in 0.01 M of Tris hydrochloride [(pH 7.4)-0.005 M EDTA-0.5% (w/v) sodium dodecyl sulfate (SDS)] on 3 wetting filters paved on a glass plate at 25 °C, and dried at 120 °C for 30 min. Then, the membranes were immersed in hybridization tubes with 10 mL of preheating prehybridization solution at 42 °C for 6 h in a shaking water bath.
Hybridization
Portions (µg) of probes were denatured with boiling water in a bath for 10 min, and then immediately placed in an ice bath for 5 min. The prehybridization solution was removed and replaced with 10 mL of hybridization solution containing 5× SSC, 45% formamide, 25 mM NaPO4 (pH 6.5), l× Denhardt solution, 250 µg of sheared denatured salmon sperm DNA per mL, 10% dextran sulfate, and 1 µg of probe DNA. The hybridization tubes were resealed and incubated for 16 h at 42 °C. The hybridization solution was removed, and the membranes were washed in 250 mL of washing buffer (250 mL of 2× SSC–1.0% (w/v) SDS) for 25 min at room temperature, and 250 mL of washing buffer for 2 min at room temperature. The membranes were then incubated in 10 mL of blocking solution (3% solution of bovine serum albumin in the same buffer) for 30 min at 42 °C, 10 mL of antibody solution for 30 min at room temperature and were rinsed briefly twice in 10 mL of washing buffer for 15 min and once in testing solution for 2–5 min at room temperature. Ten mL of color solution was added into the tubes in darkroom and rinsed with water and analyzed by scanning.
Metabolomics analysis of the serum
Supernatant preparation
The blood was immediately centrifuged at 14,000 rpm under 4 °C for 10 min, after which plasma samples were separated and stored at −80 °C until analysis. The plasma samples were prepared for the metabolomics analysis. First, the samples were thawed at room temperature before preparation. Methanol (800 µL) was then added into each sample (200 µL) to precipitate protein. Afterwards, each mixture was vortexed for 30 s, left static for 10 min at −20 °C, and then centrifuged at 14,000 rpm for 10 min at 4 °C. The supernatant (800 µL) was evaporated to dryness under a vacuum. The residue was then re-dissolved in 200 µL of methanol, and the mixture was vortexed for 1 min and centrifuged at 14,000 rpm for 10 min under 4 °C. The supernatant was retained for further analysis.
UPLC-QTOF-MS/MS-based metabolomics analysis
UPLC analysis
An ultra-performance liquid chromatograph (UPLC) analysis was performed on a Waters ACQUITY UPLC™ system (Waters Corporation, Milford, MA, USA), equipped with a binary solvent delivery system. The separation was performed with the Waters ACQUITY HSS T3 column. The flow phase was A: 0.1% formic acid, and B: acetonitrile containing 0.1% formic acid. The elution conditions of UPLC were as follows: 5% B (0–1 min), 5% to 95% B (1–10 min), 95% B (10–12 min), 95% to 5% B (12–13 min), and 5% B (13–15 min). The flow rate was set to 0.4 mL/min. The column and auto-sampler were maintained at 35 and 10 °C, respectively. The injection volume was set at 2 µL.
Mass spectrometry
A mass spectrometry was performed using a Waters Synaptic G2-SQ-TOF system (Waters MS Technologies, Manchester, UK). The flow rate of nitrogen was set at 800 L/h. The temperature of ion source was 120 °C. The capillary and cone voltages were set at 2,500 V and 40 V, respectively. The energies for collision-induced dissociations were 6 V for the precursor ion and 40–60 V for fragmentation information, respectively. The mass spectrometer and UPLC system were controlled by Mass Lynx v4.1 software. All data were acquired using Lock Spray to ensure mass accuracy and reproducibility. Mass spectrometric analysis was carried out in both positive and negative ion modes. The molecular masses of the precursor ion and of product ions were accurately determined with leucine enkephalin (m/z 554.2615) in positive mode and with leucine enkephalin (m/z 556.2771) in negative mode; the infusion flow rate was 5 L/min.
Method validation QC plasma sample preparation
The method validation of plasma sample was prepared by equally mixing each of the plasma samples (100 µL). Six QC samples (1 run) and three QC samples (3 runs) were determined in 2 consecutive days to evaluate intra-day precision and inter-day precision. The variations were evaluated by the relative standard deviation (RSDs) of peak areas of selected chromatographic peaks. The stability of the QC samples was investigated under room temperature for 8 h with 3 cycles of freezing (−80 °C) and thawing (room temperature) and under room temperature for 2, 4, 6, 8, 10 and 12 h, respectively. The data was statistically processed by principal component analysis (PCA) to determine the analytical variability over the whole run of each batch.
Multivariate statistical analysis
The original data was processed by Mass Lynx software (version 4.1, Waters Corporation, Milford, USA). The method parameters of data collection were set as follows: retention time range, 1.6–14 min; mass range, 100–1,500 Da; and mass tolerance, 15.0 ppm. For the peak integration, the peak width at 5% height was 1.00 s, the peak-to-peak base line noise was 0.00, and the noise baseline level was set at 6.00.
All of the data were normalized as relative intensity to the summed total ion intensity per chromatogram, and these data were further analyzed by PCA and orthogonal partial least square discrimination analysis (OPLS-DA). The variables with variable importance in the projection (VIP) >5 were considered to be influential for the separation of samples in the score plots generated from the OPLS-DA. MetaboAnalyst 3.0 software was performed to determine the signaling pathway with an impact value above 0.10. The experimental data were presented as mean ± standard deviation.
Statistical analysis
All experimental data are presented as mean ± standard deviation (). Data were analyzed using the Student’s t-test between 2 groups or a one-way analysis of variance among multiple groups. A P<0.05 was considered to be statistically significant, and a P<0.01 was considered to be very statistically significant. Data were calculated using SPSS17.0.
Results
Network pharmacology of LCF: overall results
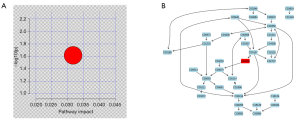
The TCMSP database was searched to identify the active components of Hedyotis diffusa, Bupleurum chinense, Sedum sarmentosum Bunge, Glycyrrhiza uralensis Fisch, Solanum nigrum L and Fructus Ligustri Lucidi in the TCMID database. A total of 133 potential active ingredients and 897 drug targets were obtained (Table 1). In addition, the “LCF—Component—Disease—Target” network analysis with Cytoscape software revealed that 133 potential active components and 185 common drug-disease targets were in LCF by deleting isolated components with no intersection (Figures 1,2). Further, a PPI network analysis with Cytoscape software was undertaken to discover the core targets. The results showed that the values of the target proteins of protein tyrosine phosphatase non-receptor type 1 (PTPN1), estrogen receptor 1(ESR1), cyclin-dependent kinase 2 (CDK2), epidermal growth factor receptor (EGFR), ATP binding cassette subfamily G member 2 (ABCG2) and cyclin-dependent kinase 1 (CDK1) were more than 3 times of the median, which indicated that these proteins were of great significance for LCF in treating CHB (Figure 1A,B,C). Further, the core genes of SRC proto-oncogene (SRC), and enhancer of zeste homolog 2 (EZH2) were obtained by core target screening based on a cluster analysis (Figure 1A,B,C,D).

Full table
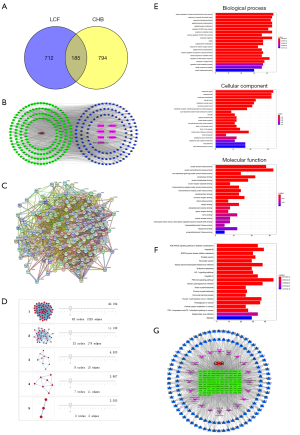

To further understand the functional implications of differentially expressed genes, 2,766 pathways were obtained from the KEGG and GO databases, including serine/threonine kinase, the response to a molecule of bacterial origin, lipopolysaccharide, and peptide-tyrosine phosphorylation. The intersection gene set revealed that 154 processes were related to molecular function, mainly including the membrane region, the membrane raft, the membrane micro domain, and the vesicle lumen. The above data showed that LCF exerted a therapeutic effect on CHB through multiple biological processes (Figure 1E). The KEGG pathways were obtained involved hepatitis B, phosphatidylinositol 3 kinase-protein kinase B (PI3K-AKT) signaling pathway, the AGE-RAGE signaling pathway in diabetic complications, and Kaposi sarcoma-associated herpes virus infection. These results indicated that the active component targets of LCF are distributed in different pathways and might play a role in the coordination of multiple pathways (Figure 1F).
Possible CHB-related pathways combined with the active components and targets of LCF were screened from the top 20 KEGG pathways to construct the disease-component-target-pathway network diagram (Figure 1G in which red represents diseases, green represents potential targets, blue represents active ingredients, and purple represents a pathway). The results showed that the active ingredients of LCF for CHB treatment were mainly metacarpi, isorhamnetin, glypallichalcone, and phaseolinisoflavan. These ingredients mainly act on targets (PTPN1, ESR1, CDK2, EGFR, ABCG2, CDK1, and PIK3CA) that are involved in signaling pathways, such as Hepatitis B, the PI3K-Akt signaling pathway, AGE-RAGE signaling pathway, and the Kaposi Sarcoma-associated herpes virus infection.
Effects of LCF on DHBV-DNA in duck serum
Effects of LCF on DHBV-DNA
We detected DHBV-DNA copies on D0, D5, D10 and P3 by dot-blot hybridization in the duck serum. The results showed that in the dot-blot hybridization maps (Figure 2A,B,C,D,E), a substantial and high expression of DHBV-DNA was measured in the model, but no difference was detected on D0 among the groups. The expression of DHBV-DNA was significantly decreased in the positive control-LAM group on D5 and D10, and the effect was better than LCF, but a rebound was detected on P3 in the positive control-LAM group, and the expression of DHBV-DNA was higher than LCF groups. In relation to each of the LCF groups, the low-dose group showed the best effect, and the expression of DHBV-DNA was obviously decreased on D10 and P3. However, there was no dose effect among the low-, medium-, and high-dose LCF groups. Thus, there was no significant correlation between the anti-HBV effect and dose.
Gray value of DHBV-DNA dots
The gray value of the DHBV-DNA dots in each group at different times are shown in Table 2. There was no difference among the groups on D0 (P>0.05). The content of DHBV-DNA was significantly decreased in the positive-LAM group on D5 and D10 compared with D0 (P<0.01) but rebounded quickly on P3. An obvious downward trend of DHBV-DNA was shown on D5, P3 in the high-dose LCF group, on P3 in the middle-dose LCF group, and on D5, D10, and P3 in the low-dose LCF group compared with the D0 of each group. However, the role of LCF in anti-HBV was not dose dependent.

Full table
The DHBV-DNA of D5, D10, and P3 in each intervention group was lower than that in the model group (P<0.01). The content of DHBV-DNA in the LCF groups on D5 and on D10 was higher than that of the LAM-positive group, but DHBV-DNA then decreased and became lower than that of LAM-positive group on P3 (P>0.05). The low dose group had the best effect of the 3 LCF groups on D5, D10, and P3, and there was a significant difference between the low-dose group and the middle-dose group. The results showed that the inhibitory effect of LAM on DHBV-DNA replication was better than LCF in 5 to 10 days, but the effect of LCF was better than LAM after 3 days’ withdrawal. Thus, LCF has a lasting effect on anti-HBV and can inhibit the rebound of DHBV-DNA.
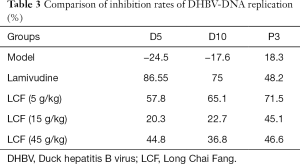
Effects of LCF on DHBV-DNA replication in duck serum
As Table 3 and Figure 2F shows, DHBV-DNA maintained a relatively stable level during the experiment process after HBV infection, but there was a slight increase on P3. The inhibition rate of DHBV-DNA in the LAM group was 86.55% on D5 and 75% on D10, but decreased to 48.2% on P3. The inhibition rate of DHBV in the 3 LCF groups increased with time and continued increasing after 3 days discontinuation without an obvious rebound phenomenon of HBV replication. The effect of the low dose of LCF on HBV was the best of the 3 groups, but there was no dose effect among the 3 groups.
Full table
Effects of LCF on the metabolomics of duck serum
PCA
To examine whether the LCF treatments alleviated the disorder of the metabolic functions caused by the virus in the ducks, PCAs were conducted based on the acquired UPLC-QTOF-MS/MS plasma data. As the score plots in Figure 3 show, the QC samples were highly superimposed in both the positive and negative ion mode, which further indicated excellent performance. In the PCA diagrams, the positive drug treatment group and all the LCF treatment groups were clustered and separated from the virus group in both ion modes. Further, after 10 days of treatment, they were significantly further away from the virus group and closer to the positive drug group. After 3 days of discontinuation, they remained isolated from the virus group and close to the positive group.
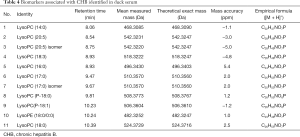
Potential biomarkers in the serum of each group
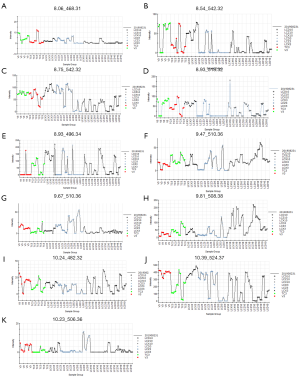
The potential plasma biomarkers associated with HBV infection were identified in this study. As Table 4 shows, all the biomarkers were associated with phospholipids catabolism. Figure 4 shows the concentration trend of 11 different metabolites in each sample. As Figure 4 shows, the content of these metabolites increased after drug administration, and there was no significant dose dependence with the increase of dose and no significant downward trend on P3. The results showed that the therapeutic effect of LCF was persistent.
Full table
Metabolic pathways
The identified metabolites information was analyzed with MetaboAnalyst 3.0 software to define which metabolic pathways were disturbed by HBV infection (Figure 5A). Figure 5B shows the phospholipid metabolism pathway and the chemical reactions involved.
Discussion
According to TCM theory and clinical experience, CHB patients are likely to be infected with a damp heat virus, which can lead to liver and gallbladder dysfunction, such as liver depression and spleen deficiency, which in turn causes disease (8,9). TCM has a number of advantages in the treatment of CHB. It improves patient symptoms, enhances liver function, and regulates immune function (9). This study suggested that the active ingredients of LCF against CHB might include metacarpi, isorhamnetin, glypallichalcone, and phaseolinisoflavan. Positive targets, such as ESR1 and PIK3CA, as well as signaling pathways, such as the Hepatitis B, PI3K-Akt and AGE-RAGE pathways, played therapeutic roles (10).
Modern pharmacological studies have shown that medicarpin has an obvious effect on human liver cancer cells. Isorhamnetin has been shown to have anti-inflammatory, anti-tumor, and anti-oxidat effects and to regulate immune function activities (11,12). Glypallichalcone and phaseolinisoflavan also have anti-inflammatory, liver-protecting, anti-oxidantion, and immunomodulatory effects (13,14). The above data showed that these components perform an important role in contributing to the actionability of LCF on CHB.
The PPI network analysis showed that LCF might treat CHB through ESR1, PIK3CA, CDK2, CDK1, EGFR and other targets. The gene polymorphism of estrogen receptor ESR1 is related to HBV infection, disease progression, and estrogen level, which can protect the liver by preventing liver fibrosis, inflammatory damage, and apoptosis (15). The PIK3CA gene is involved in cell apoptosis and tumor invasion (16). CDK2 and CDK1 are important cell cycle regulators that are involved in liver cell growth and apoptosis, which are closely related to liver inflammation (17,18). EGFR is critical in mediating HBV internalization, and it drives NTP-bound HBV to translocation from the cell surface to the endoplasmic reticulum, which leads to virus infection (19)
HBV infection is a major factor that causes chronic liver disease, fibrosis, cirrhosis, and ultimately liver cancer (20). Antiviral therapy is a first-line treatment (21). The PI3K-Akt signaling pathway is an important signal transduction pathway for HBV infection and carcinogenesis, and can promote cell growth proliferation, inhibit apoptosis, and inhibit tumor formation (22,23). Further, the AGE-RAGE signaling pathway activation can release a large number of adhesion molecules, growth factors, pro-inflammatory cytokines (such as interleukin-6, interleukin-8, and tumor necrosis factor-α), and can eventually lead to chronic cell activation and tissue damage (21).
We also investigated the effects of LCF on DHBV in a duck model (24). We found that LCF had a stronger effect on anti-HBV-DNA replication than LAM. There was no obvious difference among the low-, medium- and high-dose LCF groups. However, the anti-DHBV-DNA effect of the low-dose group was the best, which may be related to the side effects of LCF.
Metabolomics analysis was conducted to investigate the regulatory effects of LCF on the metabolic abnormality of HBV-infected ducks. We found 11 important metabolites in duck serum that mainly involved the PC and PE categories, including the LysoPC (14:0/0:0), LysoPC (16:0/0:0), LysoPC (17:0/0:0), LysoPC (P-18:0), LysoPC (P-18:1), LysoPC (18:0), LysoPC (20:5/0:0), LysoPE (18:0), LysoPC (18:3/0:0), LysoPC (20:5), and LysoPC (17:0) isomers. The decrease in the phosphatidylcholine (PC) level of them was the main mechanism of chronic liver disease progression. Research has shown that HBV infection may induce changes in the composition of PCs in HBV-infected mouse livers (25). The supplementation of exogenous choline can regulate the balance of oxidative stress in the liver, reduce liver inflammation, and improve mitochondrial energy utilization (26-28); thus phosphatidylethanolamine is also closely related to inflammation and liver fibrosis (29).
In conclusion, this study showed that LCF had an obvious effect on DHBV by inhibiting the DHBV-DNA replication rebound. Metabolites may repair liver cells through the phospholipid metabolic pathway, which could contribute to the therapeutic effects of LCF. Overall, this study revealed the molecular mechanisms and clinical application of LCF against DHBV and provides new ideas for further in-depth research.
Acknowledgments
Funding: This study was supported by the third batch of academic experience inheritance work of old traditional Chinese medicine experts in Jiangsu Province (20SSC007).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-1923
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-1923
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-1923). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Experiments were performed under a project license (No.: aEWC-20181205-65) granted by ethics committee of Jiangsu Province Hospital on Integration of Chinese and Western Medicine, in compliance with Jiangsu Province Hospital on Integration of Chinese and Western Medicine guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Makvandi M. Update on occult hepatitis B virus infection. World J Gastroenterol 2016;22:8720-34. [Crossref] [PubMed]
- Pollicino T, Caminiti G. HBV-Integration Studies in the Clinic: Role in the Natural History of Infection. Viruses 2021;13:368. [Crossref] [PubMed]
- Cui Y, Jia J. Update on epidemiology of hepatitis B and C in China. J Gastroenterol Hepatol 2013;28:7-10. [Crossref] [PubMed]
- Seto WK, Lo YR, Pawlotsky JM, et al. Chronic hepatitis B virus infection. Lancet 2018;392:2313-24. [Crossref] [PubMed]
- Wang FS, Fan JG, Zhang Z, et al. The global burden of liver disease: the major impact of China. Hepatology 2014;60:2099-108. [Crossref] [PubMed]
- European Association for the Study of the Liver. Electronic address eee, European Association for the Study of the L. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol 2017;67:370-98. [Crossref]
- Chen Y, Zhu J. Anti-HBV effect of individual traditional Chinese herbal medicine in vitro and in vivo: an analytic review. J Viral Hepat 2013;20:445-52. [Crossref] [PubMed]
- Lu Y, Fang Z, Zeng T, et al. Chronic hepatitis B: dynamic change in Traditional Chinese Medicine syndrome by dynamic network biomarkers. Chin Med 2019;14:52. [Crossref] [PubMed]
- Wang G, Zhang L, Bonkovsky HL. Chinese medicine for treatment of chronic hepatitis B. Chin J Integr Med 2012;18:253-5. [Crossref] [PubMed]
- Ghany M, Liang TJ. Drug targets and molecular mechanisms of drug resistance in chronic hepatitis B. Gastroenterology 2007;132:1574-85. [Crossref] [PubMed]
- Lu X, Liu T, Chen K, et al. Isorhamnetin: A hepatoprotective flavonoid inhibits apoptosis and autophagy via P38/PPAR-alpha pathway in mice. Biomed Pharmacother 2018;103:800-11. [Crossref] [PubMed]
- Ganbold M, Owada Y, Ozawa Y, et al. Isorhamnetin Alleviates Steatosis and Fibrosis in Mice with Nonalcoholic Steatohepatitis. Sci Rep 2019;9:16210. [Crossref] [PubMed]
- Fu H, Zhang Y, Wang X, et al. Synthesis and anti-tumor activity of novel aminomethylated derivatives of isoliquiritigenin. Molecules 2014;19:17715-26. [Crossref] [PubMed]
- Koo J, Cabarcas-Petroski S, Petrie JL, et al. Induction of proto-oncogene BRF2 in breast cancer cells by the dietary soybean isoflavone daidzein. BMC Cancer 2015;15:905. [Crossref] [PubMed]
- Stroffolini T, Esvan R, Biliotti E, et al. Gender differences in chronic HBsAg carriers in Italy: Evidence for the independent role of male sex in severity of liver disease. J Med Virol 2015;87:1899-903. [Crossref] [PubMed]
- Wang C, Che L, Hu J, et al. Activated mutant forms of PIK3CA cooperate with RasV12 or c-Met to induce liver tumour formation in mice via AKT2/mTORC1 cascade. Liver Int 2016;36:1176-86. [Crossref] [PubMed]
- Singh AK, Swarnalatha M, Kumar V. c-ETS1 facilitates G1/S-phase transition by up-regulating cyclin E and CDK2 genes and cooperates with hepatitis B virus X protein for their deregulation. J Biol Chem 2011;286:21961-70. [Crossref] [PubMed]
- Cheng P, Li Y, Yang L, et al. Hepatitis B virus X protein (HBx) induces G2/M arrest and apoptosis through sustained activation of cyclin B1-CDK1 kinase. Oncol Rep 2009;22:1101-7. [PubMed]
- Iwamoto M, Saso W, Nishioka K, et al. The machinery for endocytosis of epidermal growth factor receptor coordinates the transport of incoming hepatitis B virus to the endosomal network. J Biol Chem 2020;295:800-7. [Crossref] [PubMed]
- Liu D, Xu T, Shi B, et al. Clinical relevance of the in situ assay for HBV DNA: a cross-sectional study in patients with chronic hepatitis B. J Clin Pathol 2020;73:813-8. [Crossref] [PubMed]
- Takkenberg RB, Jansen L, de Niet A, et al. Baseline hepatitis B surface antigen (HBsAg) as predictor of sustained HBsAg loss in chronic hepatitis B patients treated with pegylated interferon-alpha2a and adefovir. Antivir Ther 2013;18:895-904. [Crossref] [PubMed]
- Guo H, Zhou T, Jiang D, et al. Regulation of hepatitis B virus replication by the phosphatidylinositol 3-kinase-akt signal transduction pathway. J Virol 2007;81:10072-80. [Crossref] [PubMed]
- Xue G, Restuccia DF, Lan Q, et al. Akt/PKB-mediated phosphorylation of Twist1 promotes tumor metastasis via mediating cross-talk between PI3K/Akt and TGF-beta signaling axes. Cancer Discov 2012;2:248-59. [Crossref] [PubMed]
- Guo WN, Zhu B, Ai L, et al. Animal models for the study of hepatitis B virus infection. Zool Res 2018;39:25-31. [PubMed]
- Park ES, Lee JH, Hong JH, et al. Phosphatidylcholine alteration identified using MALDI imaging MS in HBV-infected mouse livers and virus-mediated regeneration defects. PLoS One 2014;9:e103955 [Crossref] [PubMed]
- van der Veen JN, Kennelly JP, Wan S, et al. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim Biophys Acta Biomembr 2017;1859:1558-72. [Crossref] [PubMed]
- Cao M, Li X, Zhang B, et al. The effect of polyene phosphatidyl choline intervention on nonalcoholic steatohepatitis and related mechanism. Am J Transl Res 2016;8:2325-30. [PubMed]
- Ling J, Chaba T, Zhu LF, et al. Hepatic ratio of phosphatidylcholine to phosphatidylethanolamine predicts survival after partial hepatectomy in mice. Hepatology 2012;55:1094-102. [Crossref] [PubMed]
- van der Veen JN, Lingrell S, Gao X, et al. Pioglitazone attenuates hepatic inflammation and fibrosis in phosphatidylethanolamine N-methyltransferase-deficient mice. Am J Physiol Gastrointest Liver Physiol 2016;310:G526-38. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)



