
Long non-coding RNA SNHG1 mediates neuronal damage in Parkinson’s disease model cells by regulating miR-216a-3p/Bcl-2-associated X protein
Introduction
Parkinson’s disease (PD) is an extremely common neurodegenerative disease, the general clinical characteristics of which include muscle stiffness, slow movement, resting tremor, and staggering (1). PD is characterized by the selective loss of substantia nigra nerve cells in the brain, resulting in a decrease in substantia nigra dopamine neurons. Despite long-term and in-depth clinical, physiological, and biochemical research on PD having been conducted, the exact cause of the pathological changes associated with the disease has remained unclear. Many factors, including environmental, age-related, and heredity factors, can contribute to the degeneration and death of dopaminergic neurons (2,3). There is still no clinically effective drug for the treatment of PD. Therefore, clinically, drugs are often used for the treatment of PD. Doctors prescribe drugs with the aims of delaying the development of the disease and maintaining the patient’s quality of life (4-6). Therefore, further clarification of the pathogenesis of PD is urgently needed.
Long non-coding RNAs (lncRNAs) are non-coding RNAs that cannot be transcribed to produce cellular protein. In recent years, an increasing number of studies have demonstrated that lncRNAs are important regulators in the pathogenesis of PD (7,8). Small nucleaolar RNA host gene 1 (SNHG1) is one of the lncRNAs that have been studied more in recent years. A number of references report that it plays an important role in lung cancer and liver cancer, participates in many signal pathways, and becomes a target site for cancer detection (9). MicroRNAs (miRNAs or miRs) are another type of non-coding RNA with a length of 18 to 24 nt that cannot be transcribed to produce cellular protein. Through transcriptional and post-transcriptional regulation, miRNAs play important roles in multiple malignant physiological cellular processes, such as oncogenesis, and participate in various cellular activities (10). Numerous studies have shown that miR-216a-3p inhibits cell proliferation and invasion as well as other cellular activities in a variety of cancer cells by regulating its target genes. Cancer cells have a high rate of apoptosis and autophagy, and miR-216a-3p has the potential to become a molecular marker in the detection of some cancers. However, whether or not miR-216a-3p plays an equally important role in the occurrence of PD has yet to be determined. Therefore, the present study explored the biological roles of lncRNA SNHG1 and miR-216a-3p in the pathogenesis of PD, as well as their relationship and mechanism in the progression of the disease. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-21-1613).
Methods
Cell culture and PD model establishment
The human neuroblastoma cell line SK-N-SH was purchased from the Shanghai Institute of Life Sciences. The SK-N-SH cells were cultured in high-sugar Dulbecco’s Modified Eagle Medium (Thermo Fisher Scientific, USA) containing working concentrations of fetal bovine serum and penicillin-streptomycin of 10% and 1%, respectively. Cell culture was carried out in an incubator with the temperature set at 37 °C and a CO2 concentration of 5%. To establish a PD cell model, cells were treated with 0.25, 0.5, or 1 mM 1-methyl-4-phenylpyridinium (MPP+) for 24 hours.
Plasmid transfection
1×105 of cells in the logarithmic growth phase were seeded on a 48-well plate and cultured until reaching 40–60% confluence. Mixtures containing 50 µL serum-free medium and 3 µL Lipofectamine 2000, and 50 µL serum-free medium and 400 ng plasmid mixture were prepared for transfection, which was the amount of one hole of the 48-well plate. Each of the 2 mixtures was mixed and stood at room temperature for 5 minutes. After that, each mixture was mixed for a second time and left to stand at room temperature for 20 minutes. The reagents were configured according to the number of holes in the experiment.
The BAX overexpression plasmid was purchased from addgene, and the plasmid name was pCEP4-HA-BAX (Plasmid # 16587). The siRNA was synthesized in Shanghai Jima Pharmaceutical Co., Ltd. (China). The siRNA specific sequences were: siSNHG1-1: target sequence + 2nt overhang: CAGCTTGTTGCTATATACCATTG; 21 nt guide (5'-3'): AUGGUAUAUAGCAACAAGCUG; 21 nt passenger (5'-3'): GCUUGUUGCUAUAUACCAUUG. siSNHG1-2: target sequence + 2 nt overhang: CTGAGCAAATAAGGTGTATAAAA: 21 nt guide (5'-3'): UUAUACACCUUAUUUGCUCAG; 21 nt passenger (5'-3'): GAGCAAAUAAGGUGUAUAAAA.
Extraction of total cell RNA and polymerase chain reaction analyses
Total RNA was extracted from SK-N-SH cells using TRIzol reagent; following extraction, the quality of the RNA was tested and quantified. Then, reverse transcription and real-time fluorescent quantitative polymerase chain reaction (qRT-PCR) were performed using the relevant kits following the manufacturers’ instructions. To detect the expression level of miR-216a-3p, due to the short lengths of miRNAs, the tailing miScript II RT Kit (Qiagen, Germany) was used for reverse transcription. To detect the mRNA expression level of Bcl-2-associated X protein (BAX), a HiScript 1st Strand cDNA Synthesis Kit (Nanjing Novezan Biotechnology Co., Ltd.) was used for reverse transcription. The primer used for miR-216a-3p detection was a matching primer designed according to the Qiagen tailing reverse transcription kit. The primers used for the detection of the SNHG1 gene and other related genes were: SNHG1-F: 5'-CTGTTCCCGTCATGAGCCTT-3'; SNHG1-R: 5'-GCAAGGCCCTGAATGAGCTA-3'; BAX-F: 5'-CATGGGCTGGACATTGGACT-3'; BAX-R: 5'-AAAGTAGGAGAGGAGGCCGT-3'; GAPDH-F: 5'-GACAGTCAGCCGCATCTTCT-3'; GAPDH-R: 5'-GCGCCCAATACGACCAAATC-3'; U6-F: 5'-CTCGCTTCGGCAGCACA-3'; U6-R: 5'-AACGCTTCACGAATTTGCGT-3'.
Western blot
First, SK-N-SH cells were collected in an Eppendorf tube. Considering the location of the protein to be detected in the cell, the cells were lysed with RIPA lysis buffer at an appropriate strength. The bicinchoninic acid assay (BCA) method was used for protein quantification. Equal amounts of protein were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After separation, the proteins were transferred to a membrane, which was blocked, and incubated with primary and then secondary antibodies. Finally, the protein bands were developed and quantified.
Cell viability experiment
To detect cell viability, a cell counting kit-8 (CCK-8) assay was performed. Briefly, cells were seeded in a 96-well culture plate at 2×104/mL for 24 hours. Then, 10 µL of CCK-8 solution was added to each well, and the cells were incubated for 2 hours. Finally, the absorbance value was measured at 450 nm with a microplate reader.
Cell apoptosis experiment
For the cell apoptosis assay, cells were digested with trypsin and centrifuged to remove the supernatant. Then, the cells were resuspended in binding buffer and stained with isothiocyanate (FITC) and propidium iodide (PI) in the dark at room temperature. Finally, according to the manufacturer’s instructions, a flow cytometer was used to detect cell apoptosis, and the percentage of apoptotic cells was recorded.
Luciferase reporter gene
psiCHECK2TM plasmids containing fragments of miR-216a-3p target sites on the three prime untranslated region (3'UTR) of SNHG1 and Bcl-2-associated X protein (BAX, including MUT) were constructed and were denoted as psiCHECK2TM-SNHG1 and psiCHECK2TM-BAX, respectively. Human embryonic kidney (HEK) 293T cells were seeded in a 24-well plate and cultured to 50–60% confluence. Anti-miR-216a-3p synthesized in Shanghai Jima Pharmaceutical Co., Ltd. (China) and the constructed related reporter gene plasmid were simultaneously transfected into the same well. The negative control (NC) group was transfected with NC (bundled with anti-miRNA synthesis) and related reporter gene plasmids. After 24 hours, the supernatant was removed, and the cell pellets were collected. Then, the cells were lysed with passive lysis buffer, and centrifuged; 20 µL of the supernatant was obtained in a 96-well white plate. After that, Firefly luciferase activity was detected using Luciferase Assay Reagent II. Stop & Glo Reagent reagents were added to inhibit luciferase activity and Renilla luciferase activity was detected.
Data analysis
Data were collected and statistically analyzed in GraphPad Prism 5.0 software, and relevant histograms were also generated using the software.
Results
Knock-down of SNHG1 affected the cell activity and apoptosis of neurons in the PD model cells
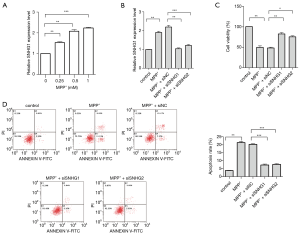
The PD cell model was constructed using MPP+ to treat the human neuroblastoma cell line SK-N-SH. Observation of the changes in SNHG1 expression before and after treatment revealed that the expression level of SNHG1 was positively correlated with the dose of MPP+ (Figure 1A). In the subsequent experiments, MPP+ was used at a concentration of 1 mM to construct an in vitro simulated PD cell model. The expression of SNHG1 was found to be significantly increased in the PD model cells. To establish whether SNHG1 expression was related to the growth of neurons in the model, we knocked out neuronal expression of SNHG1. After SNHG1 knock-down, the cell activity of the PD model cells increased, while the apoptotic rate decreased (Figure 1B,C,D).
The interaction between SNHG1 and miR-216a-3p
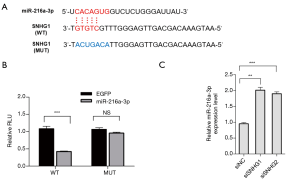
To further explore the role of SNHG1 in neurons in the PD model, we screened miR-216a-3p as the target of SNHG1 in the database (TarBase and miRWalk) (Figure 2A). The luciferase reporter gene experiment was used to verify the interaction between SNHG1 and miR-216a-3p (Figure 2B). The results of qRT-PCR showed that the expression of miR-216a-3p was upregulated after knock-down of SNHG1 (Figure 2C). Together, the above experimental results suggested that SNHG1 could directly bind to miR-216a-3p to negatively regulate its expression level.
miR-216a-3p alleviated neuronal damage in the PD cell model after SNHG1 knock-down
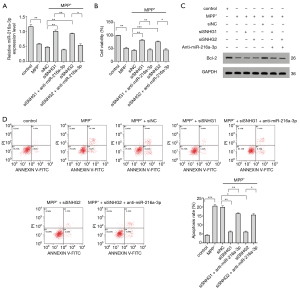
After confirming the interaction and negative regulatory relationship between SNHG1 and miR-216a-3p, we attempted to establish whether miR-216a-3p was involved in the effect exerted by SNHG1 on neuronal cell activity and apoptosis. In the MPP+-treated PD model cells, the expression of miR-216a-3p was downregulated; however, after knock-down of SNHG1, the expression level of miR-216a-3p increased (Figure 3A). However, in cells transfected with anti-miR-216a-3p, miR-216a-3p expression was low even after SNHG1 knock-down. After knock-down of SNHG1, cell activity in the PD model cells increased, while the decline in the apoptotic rate was alleviated (Figure 3B,C,D).
BAX is a target of miR-216a-3p
Combining the predicted sequence of miR-216a-3p with the existing database, we predicted BAX to be a target of miR-216a-3p in PD (Figure 4A). A luciferase reporter gene experiment was used to verify the relationship between miR-216a-3p and BAX, and the experimental results confirmed that BAX was indeed a target of miR-216a-3p in PD (Figure 4B).

BAX reversed the increase in neuronal activity and the decrease in apoptosis in PD model cells when miR-216a-3p was overexpressed
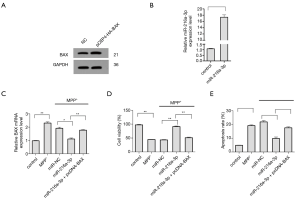
The results of our experiments so far had shown that the overexpression of miR-216a-3p changed the cell conditions in the PD cell model caused by SNHG1 knock-down. However, whether the mitigation effect of miR-216a-3p could be achieved by regulating its target BAX was unclear. Therefore, we sought to verify this in our next experiment. First, the effective expression of pCEP4-HA-BAX and miR-216a-3p mimics after transfection into cells was verified (Figure 5A,B). Then, BAX was overexpressed in cells also overexpressing miR-216a-3p. The results of qRT-pCR showed that in these cells, the expression of BAX was increased, the cell viability was reduced, and the apoptosis rate was increased compared to cells only overexpressing miR-216a-3p (Figure 5C,D,E).
Discussion
PD, also known as tremor paralysis, is an extremely common neurodegenerative disease, which mostly affects older people (1-4). Since PD was first discovered, many clinical and biological studies have been conducted; however, the exact cause of the pathological changes in PD is still unclear. Numerous researches have shown that PD occurrence is related to environmental and genetic factors working together. Therefore, from the perspective of genetic molecular biology, the transcriptional and translational regulation of related genes or genetic molecules plays an important role in the occurrence and development of PD.
Existing studies have shown that lncRNAs are involved in the development of the brain and in the maintenance of neuronal function and life activities. For instance, lncRNA UCA1 can aggravate the damage to dopaminergic neurons in rats with PD by activating the PI3K/Akt signaling pathway, causing inflammation and oxidative stress, while down-regulating lncRNA HOTAIR can suppress the development of PD by regulating the expression of LRRK2 (11,12). Therefore, lncRNAs have gradually become the main biomarkers in exploration of the pathogenesis and treatment strategies of PD.
Apoptosis is an important cellular life process which is responsible for maintaining homeostasis in the brain. Abnormalities in apoptosis have a particular impact on the occurrence and development of PD and are considered to be an important sign of PD-related neuronal degradation (13). Therefore, regulating cell apoptosis may provide new ideas for the effective clinical treatment of PD.
In recent years, studies have shown that the lncRNA SNHG1 may be involved in the pathogenesis of PD (14,15). In the present study, we treated human neuroblastoma cell lines with MPP+ to construct a PD cell model, and further studied the role of SNHG1 in the development of PD. Our results demonstrated that the expression of SNHG1 was increased in the PD model cells. Knock-down of SNHG1 inhibited cell viability in the model cells while increasing the apoptotic rate. Through sequence alignment, we predicted the miRNA target of SNHG1 and found that SNHG1 can be used as an auxiliary device for miR-216a-3p; thus, miR-216a-3p became a target molecule of study in this research. In this study, the expression of miR-216a-3p was reduced in PD model cells, and re-expression could reverse the high apoptosis rate and inhibition of cell vitality observed in the PD model. These findings indicate that SNHG1 is an upstream molecule that can adsorb and negatively regulate miR-216a-3p, and that miR-216a-3p can attenuate the effect of SNHG1 knock-down on neurons.
To further explore the specific network roles of SNHG1 and miR-216a-3p in the development of PD, we turned our attention to BAX, as 1 of the targets of miR-216a-3p. BAX, a member of the BAL-2 family, is a regulator of cell apoptosis that participates in various activity relating to neuronal injury, which can lead to neuronal damage and cell apoptosis (16-19). However, there are few studies on its relationship with PD occurrence and development. The results of our research showed that the overexpression of BAX blocked the neuronal damage by miR-216a-3p. At the same time, SNHG1 acted as a coordinator by mediating the regulation of BAX by miR-216a-3p, thereby affecting the activity and apoptosis of neurons in the PD model. These interactions form a SNHG1/miR-216a-3p/BAX molecular regulatory network, which plays a role in the pathogenesis of PD.
In conclusion, the present study has shown that SNHG1 interacts with miR-216a-3p and that BAX is a target of miR-216a-3p in PD. The expression of miR-216a-3p increases in PD model cells following SNHG1 knock-down, while the expression of BAX decreases with miR-216a-3p overexpression. Therefore, the expression of BAX in PD model cells decreases after SNHG1 knock-down. We have also confirmed that SNHG1 can indirectly regulate the expression of BAX via miR-216a-3p, thereby affecting neuronal damage in PD model cells. Through this study, we have discovered the SNHG1/miR-216a-3p/BAX molecular regulatory network in the pathogenesis of PD, which provides us with strong evidence for further research and treatment of PD. This study has only obtained relevant conclusions in the Parkinson's model of cells, but this molecular regulatory network in animal models is still to be studied. Therefore, there is still a long way to go for the treatment of PD caused by the network-related factors.
Acknowledgments
Funding: This work was supported by Public Welfare Project of Zhejiang Science and Technology Department (2017C37150, 2017C33114) and Zhejiang Medical Science and Technology General Project (2018KY033).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-1613
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-1613
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-1613). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dauer W, Przedborski S. Parkinson's disease: mechanisms and models. Neuron 2003;39:889-909. [Crossref] [PubMed]
- Reid WG, Hely MA, Morris JG, et al. Dementia in Parkinson's disease: a 20-year neuropsychological study (Sydney Multicentre Study). J Neurol Neurosurg Psychiatry 2011;82:1033-7. [Crossref] [PubMed]
- Irwin DJ, Lee VM, Trojanowski JQ. Parkinson's disease dementia: convergence of α-synuclein, tau and amyloid-β pathologies. Nat Rev Neurosci 2013;14:626-36. [Crossref] [PubMed]
- Meng YH, Wang PP, Song YX, et al. Cholinesterase inhibitors and memantine for Parkinson's disease dementia and Lewy body dementia: A meta-analysis. Exp Ther Med 2019;17:1611-24. [PubMed]
- Sulzer P, Liebig L, Csoti I, et al. A time-efficient screening tool for activities of daily living functions in Parkinson's disease dementia. J Clin Exp Neuropsychol 2020;42:867-79. [Crossref] [PubMed]
- Garcia-Ptacek S, Kramberger MG. Parkinson Disease and Dementia. J Geriatr Psychiatry Neurol 2016;29:261-70. [Crossref] [PubMed]
- Gendron J, Colace-Sauty C, Beaume N, et al. Long non-coding RNA repertoire and open chromatin regions constitute midbrain dopaminergic neuron - specific molecular signatures. Sci Rep 2019;9:1409. [Crossref] [PubMed]
- Elkouris M, Kouroupi G, Vourvoukelis A, et al. Long Non-coding RNAs Associated With Neurodegeneration-Linked Genes Are Reduced in Parkinson's Disease Patients. Front Cell Neurosci 2019;13:58. [Crossref] [PubMed]
- Thin KZ, Tu JC, Raveendran S. Long non-coding SNHG1 in cancer. Clin Chim Acta 2019;494:38-47. [Crossref] [PubMed]
- O'Brien J, Hayder H, Zayed Y, et al. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne) 2018;9:402. [Crossref] [PubMed]
- Cai L, Tu L, Li T, et al. Downregulation of lncRNA UCA1 ameliorates the damage of dopaminergic neurons, reduces oxidative stress and inflammation in Parkinson's disease through the inhibition of the PI3K/Akt signaling pathway. Int Immunopharmacol 2019;75:105734 [Crossref] [PubMed]
- Mu G, Liu Q, Wu S, et al. Long noncoding RNA HAGLROS promotes the process of mantle cell lymphoma by regulating miR-100/ATG5 axis and involving in PI3K/AKT/mTOR signal. Artif Cells Nanomed Biotechnol 2019;47:3649-56. [Crossref] [PubMed]
- Tatton WG, Chalmers-Redman R, Brown D, et al. Apoptosis in Parkinson's disease: signals for neuronal degradation. Ann Neurol 2003;53:S61-70; discussion S70-2. [Crossref] [PubMed]
- Qian C, Ye Y, Mao H, et al. Downregulated lncRNA-SNHG1 enhances autophagy and prevents cell death through the miR-221/222 /p27/mTOR pathway in Parkinson's disease. Exp Cell Res 2019;384:111614 [Crossref] [PubMed]
- Xie N, Qi J, Li S, et al. Upregulated lncRNA small nucleolar RNA host gene 1 promotes 1-methyl-4-phenylpyridinium ion-induced cytotoxicity and reactive oxygen species production through miR-15b-5p/GSK3β axis in human dopaminergic SH-SY5Y cells. J Cell Biochem 2019;120:5790-5801. [Crossref] [PubMed]
- Miyashita T, Krajewski S, Krajewska M, et al. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene 1994;9:1799-805. [PubMed]
- Yang X, Zhang M, Wei M, et al. MicroRNA-216a inhibits neuronal apoptosis in a cellular Parkinson's disease model by targeting Bax. Metab Brain Dis 2020;35:627-35. [Crossref] [PubMed]
- Tatton NA. Increased caspase 3 and Bax immunoreactivity accompany nuclear GAPDH translocation and neuronal apoptosis in Parkinson's disease. Exp Neurol 2000;166:29-43. [Crossref] [PubMed]
- Venkatesh Gobi V, Rajasankar S, Swaminathan Johnson WM, et al. Antiapoptotic role of Agaricus blazei extract in rodent model of Parkinson's disease. Front Biosci (Elite Ed) 2019;11:12-9. [Crossref] [PubMed]


