
Methylated biomarkers for breast cancer identified through public database analysis and plasma target capture sequencing
Introduction
Breast cancer is the most common malignancy and the leading cause of death in women worldwide. It is widely accepted that methylation occurs early in the development of cancers, including breast cancer (1). DNA methylation occurs when DNA is modified by the addition of a methyl group to the 5' position of a cytosine preceding a guanosine (CpG). CpGs are often found at high densities in ‘CpG islands’, particularly within the promoter regions of genes. Hypermethylation of CpG islands can result in the transcriptional silencing of tumour suppressor genes in cancer, whereas CpG hypomethylation may lead to oncogene activation, and which may also promote the occurrence and development of breast cancer. Aberrant promoter methylation levels of tumor related pathway genes (such as DNA damage repair genes, apoptosis related genes, drug transport and drug metabolism genes) may affect the sensitivity of tumor cells to chemotherapy drugs. Given that the DNA methylation pattern of carcinomas may differ from that of normal tissue, it is possible that certain methylated genes could be used as biomarkers in non-invasive blood-based assays, thereby enhancing diagnostic capabilities. For example, the methylation of the promoter region in circulating cell-free DNA (cfDNA) is under review by the Food and Drug Administration (FDA) as a biomarker for clinical use (2). A few studies have also demonstrated that methylated biomarkers in peripheral blood, such as BRCA1 (3), GSTP1 (4,5), RASSF1A (5,6), RARβ2 (7), and APC (4,7), can improve breast cancer diagnosis. However, the feasibility of these biomarkers for clinical use remains unknown, and a reliable method to identify clinically valuable methylated biomarkers is not currently available. The development of high-information content assays focused on abnormalities in DNA methylation, as well as the establishment of various bioinformatics databases has helped to better understand this inherent mechanism (8).
In this study, we explored public databases and conducted methylation-specific whole-gene sequencing in an attempt to explain the mechanism of gene methylation in breast cancer and to search for biomarkers that would allow for early, non-invasive diagnosis of breast cancer.
We present the following article in accordance with the STARD reporting checklist (available at http://dx.doi.org/10.21037/atm-21-1128).
Methods
Acquisition of TCGA data and Blueprint Epigenome data
Methylation data of breast cancer tissue and normal tissue on the Infinium® HumanMethylation450 BeadChip (Illumina) platform were downloaded from The Cancer Genome Atlas (TCGA) in June 2017. HM450K methylation data included 96 normal and 791 tumour samples.
Whole-genome bisulfite sequencing data of white blood cells (WBCs) were acquired from the Blueprint Epigenome Database in June 2017. The sequencing data of 26 samples were obtained, which included T cells (n=8), B cells (n=4), neutrophils (n=6), and monocytes (n=8).
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Methyl-capture sequencing of circulating cell-free DNA
For the methylation analysis of cfDNA, 10 female patients with early-stage breast cancer and 10 healthy female volunteers from Nanfang Hospital of Southern Medical University were enrolled in this study between March 2018 and July 2018. Patients in the experimental group were matched with healthy individuals in the control group in terms of age, body mass index (BMI), and menstrual status. None of the patients had undergone chemotherapy or radiotherapy before surgery. The breast cancer diagnosis of the experimental group and healthy status of the control group were confirmed by core needle biopsy. The clinicopathological information of all patients is listed in Table 1. Whole peripheral blood (10 mL for each participant) was collected from the ante-cubital fossa of all patients with operable breast cancer before tumour excision was carried out. This study was approved by the Ethics Review Board of Nanfang Hospital of Southern Medical University, and informed written consent was obtained from all participants.
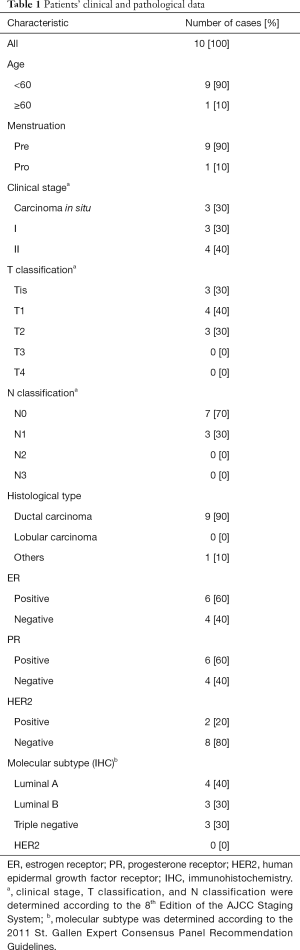
Full table
Plasma was separated by centrifugation (1,600 ×g) within 8 h of blood sample collection and kept at −80 °C until further processing. Sample processing was conducted by CapitalBio Genomics Co., Ltd. (Dongguan, China). CfDNA was extracted using the Mapure Circulating DNA Kit (Magen, Shanghai China). Ten nanograms of cfDNA samples were then used for whole-genome bisulfite sequencing library construction using the Acegen Ultralow Bisulfite-Seq Library Prep Kit (Acegen, Shenzhen China) according to the manufacturer’s instructions.
The liquid hybridization capture procedure is described below. Briefly, 125 ng of DNA from each of the eight adapter-ligated libraries were pooled together. One hundred micrograms of bisulfite capture enhancer and 1 nmol of adapter complementary DNA oligos were added to the above solution. The mixture was subsequently dried using SpeedVac (Eppendorf, Shanghai China) at 60 °C, and denatured at 95 °C for 10 min at a final reaction volume of 10.5 µL (7.5 µL 2X Hybridization Buffer and 3 µL Hybridization Component A). After centrifugation, 4.5 µL of designed probes were added, and the mixture was hybridized in a thermal cycler at 47 °C for 72 h with the lid heated at 57 °C. After hybridization, the biotin-labelled DNA probes were captured using 100 µL Dynabeads M-280 Streptavidin (Invitrogen, Shanghai China), which had been pre-washed twice with 400 µL Streptavidin Dynabead Binging and Washing Buffer. The capture program was incubated at 47 °C for 45 min in a thermal cycler with the lid heated at 57 °C, and the mixture was vortexed for 3 s every 15 minutes during the incubation period. Unbound fractions were discarded, and DNA-probe-bead complexes were collected and washed once with 100 µL 1X Wash Buffer I prewarmed to 47 °C, before being washed twice with 400 µL 1X Stringent Wash Buffer I prewarmed to 47 °C for 5 min (X represents the volume of DNA and varies with the concentration of DNA). After the supernatant was discarded, the collected bead complexes were consecutively washed again with 200 µL 1X Wash Buffer I, 200 µL 1X Wash Buffer II, and 200 µL 1X Wash Buffer III. All hybridization and wash buffers were purchased from Roche NimbleGen (SeqCap EZ Hybridization and Wash Kits). Finally, the captured DNA was eluted with 50 µL 10 M and incubated at room temperature for 10 min. The supernatant was transferred into a new tube and neutralized with 50 µL 10 M efore being purified using MiniElute Purification Kit (Qiagen, Shanghai China).
To effectively capture the related methylated regions of plasma cfDNA, we applied SeqCap Epi CpGiant Probes (Roche, Shanghai China), which could interrogate more than 5.5 million methylation sites with a total capture size of 80.5 MB, to capture target-selected genomic regions from prepared sequence libraries of bisulfite-treated cfDNA, while the construction of each library only required about 10 ng of initial cfDNA (9,10). The captured library concentrations were measured using the Qubit 2.0 Fluorometer and quantitative PCR, and the insert size was checked with an Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). The resultant libraries were sequenced using Illumina Hiseq X Ten system according to the manufacturer’s instructions. Raw sequencing data were processed using the Illumina base-calling pipeline. Low-quality reads [which contained more than 30% ‘N’s or over 10% of the sequence with low quality value (quality value <20) per read] were excluded from the data analysis. The clean reads were aligned to the human reference genome (hg19) using rogram (11).
BSP-HiSeq of circulating cell-free DNA
Bisulfite-converted cfDNA samples were subjected to Multi-BSP-HiSeq steps. Multiplex PCR [Platinum® Multiplex PCR Master Mix Kit (LifeTechnologies, Carlsbad, CA, USA)] was performed in a 10-µL reaction volume consisting of 5-µL 2× Master Mix Buffer, 4.5-µL bisulfite-converted cfDNA sample (15 ng/µL), and 0.5-µL primer mix of the six candidate genes (50 nM/per primer). The thermocycler setting was 95 °C for 2 min, followed by 15 cycles of 95 °C for 15 s and 58 °C for 4 min, and then a final extension of 3 min at 72 °C. The BSP primers of all six CpGs were designed using MethPrimer (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi). The products of multiplex-PCR were subjected to library construction with end-polishing, A-tailing, adapter ligation, and five-cycle PCR to generate barcoded libraries. Barcoded libraries from different samples were then pooled together equally and sequenced using Illumina Hiseq X Ten system (Illumina,9885 Towne Centre Drive San Diego, CA 92121 USA) according to the manufacturer’s instructions.
Statistical analysis
Data analysis was performed using bioinformatics programs written in Perl and R. A P value ≤0.001 was considered statistically significant.
Variance analysis was used to evaluate the difference of methylated cytosine-phosphate-guanine (CpG) sites between breast tumour and normal samples [false discovery rate (FDR) <0.001, |Delta Beta| >0.2]. P values were corrected using the Bonferroni method, and Tukey’s honest significant difference test was applied. The Kyoto Encyclopaedia of Genes and Genomes (KEGG) was utilized for the annotation of signalling pathways using the online tool (https://david.ncifcrf.gov/).
Raw sequencing data were pre-treated with Bioconductor minfi package (http://bioconductor.org/packages/release/bioc/html/minfi.html). The filtered data were used for group comparison analysis of cfDNA methylations between patients and healthy individuals using the Student’s t-test (P value <0.05, |Delta Beta| >0.2) for each CpG site with at least 5X sequencing depth in more than five samples within each group. The beta-values were used for unsupervised hierarchical clustering analysis of the methylation status of identified CpG sites. DAVID was also used for functional annotation of the related genes.
To further identify methylated CpGs as biomarkers for breast cancer, we compared the aberrant methylation in breast cancer tissues with the methylation data of WBCs acquired from the Blueprint Epigenome Database. The Pearson correlation coefficient calculation was used for Scatter plot analysis between breast cancer tissue, normal breast tissue, and WBCs. The Student’s t-test and Benjamin correction method were used to compare the differentially methylated CpGs (DMCs) (FDR <0.001, |Delta Beta| >0.2) between breast tumour samples and WBCs. Lastly, density analysis was performed for these wbc-DMCs in breast cancer tissue, normal breast tissue, and WBCs.
The sequencing data will be uploaded to SRA database soon.
Results
Differential DNA methylation in breast cancer and normal tissue based on TCGA
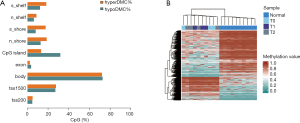
To investigate aberrant DNA methylation in breast cancer, we compared breast cancer and normal breast tissue samples in TCGA. A total of 23,088 DMCs sites were identified between breast cancer samples and normal breast tissue samples. Of the breast tumour DMCs (T-DMCs), hypermethylation was detected in 13,387 sites (57.98%), with the majority (78.4%, 10,449) in the CpG island and flanking shores (CpG island, 7,289; shores, 3,160). Only 3.9% (370) of hypomethylated T-DMCs (hypoT-DMCs) were located in the CpG island.
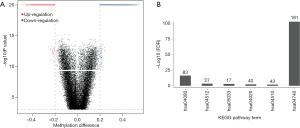
To further understand the potential role of these aberrant methylations in breast cancer, we applied functional annotation analysis of genes with DMCs in 1,500 regions flanking transcription start sites using DAVID. Our results indicated that the hypomethylated site-associated genes were enriched in the olfactory transduction pathway (FDR =7.06E-104). Hypermethylated site-associated genes were enriched in five pathways, including the neuroactive ligand-receptor interaction (FDR =4.02E-17), the extracellular matrix (ECM)-receptor interaction (FDR =3.73E-04), nicotine addiction (FDR =9.75E-04), the calcium signalling pathway (FDR =4.31E-03), and focal adhesion (FDR =1.08E-02) (Figure 1).
Methylation analysis of plasma cfDNA for breast cancer diagnosis
Plasma samples of 10 breast cancer patients and 10 healthy individuals from Nanfang Hospital of Southern Medical University were analysed using target capture sequencing, with an average of 14.0 GB of raw sequence data generated for each sample. 75.36% of the clean reads (after removal of low-quality reads) were mapped onto the hg19 reference genome, resulting in an average sequencing depth of 9.47X at 7.90 million CpG sites. Bisulfite conversion efficiency (>98.7%) was validated by the methylation levels, since methylation does not occur at CHH sites in mature animal cells (12,13). However, only 30.0% of the mapped reads were aligned to the target regions, indicating that efficiency of the capture reactions might have been affected by short fragment length and low abundance of cfDNA. Group comparison analysis of cfDNA methylations between patients and healthy individuals identified a total of 1,453 DMCs of cfDNA (cf-DMCs). Of these cf-DMCs in breast cancer patients, hypomethylation was detected at 937 sites (64.49%), mostly in the CpG island and gene body. Although the breast cancer samples used in this study came from patients with different stages of breast cancer, unsupervised hierarchical clustering analysis of the methylation status of cf-DMCs could cluster samples of the same group together, indicating that the methylation of these sites could be used as a biomarker for breast cancer diagnosis (Figure 2).
Functional annotation analysis using DAVID was also carried out for all 146 genes with hyperDMCs and the 204 genes with hypoDMCs in 1,500-bp regions flanking transcription start sites (14,15). No significant enrichment term was observed.
Plasma cfDNA methylation for early breast cancer diagnosis
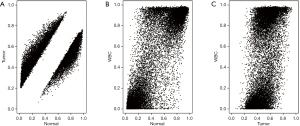
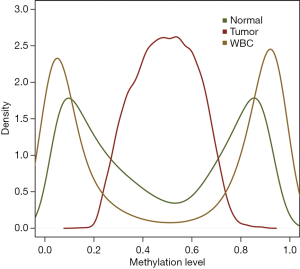
To further identify methylated CpGs for breast cancer diagnosis, we compared the aberrant methylation in breast cancer with the methylation data of WBCs. Scatter plot analysis showed that aberrant methylated CpGs in breast cancer was highly correlated with those in WBCs and normal breast tissue (Pearson’s r=0.86, P=0) (Figure 3). In addition, the comparative analysis of the DMCs of breast cancer samples and WBCs generated a total of 14,003 (wbc-DMCs). Density analysis of these wbc-DMCs further illustrated a beta-distribution in the methylations of WBCs and normal breast tissue, with 83.13% (11,640) of wbc-DMCs methylation similar to (|Delta Beta| <0.2) that in normal breast tissue (Figure 4). Various DNA methylation profiling studies have identified candidate biomarkers for breast cancer diagnosis (16-19), and our results supported the importance of these methylation markers in plasma cfDNA for cancer screening.
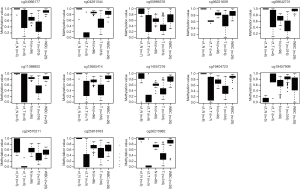
Our research showed that methylation levels in 13 CpG sites (cg04066177, cg04281344, cg05995576, cg06221609, cg08642731, cg11388802, cg12665414, cg14557216, cg19404723, cg19457909, cg24570211, cg25818763, and cg26215982) were highly comparable in the malignant tissue DNA and cfDNA, but differed significantly from those of adjacent normal tissue DNA and cfDNA, as well as leukocyte DNA (Figure 5). These CpG sites were annotated to 10 protein-coding genes, including FABP9, LCE1A, CACNA1E, CPEB4, DNAI1, DLGAP2, BCAS3, CTNNA2, DCC, and GALNT8.
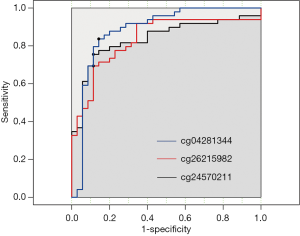
To identify whether CpGs could be used as biomarkers for early-stage breast cancer diagnosis, we further analysed the methylations of three CpGs (cg04281344, cg24570211, and cg26215982), which were hypomethylated in cfDNA from cancer patients, in an independent patient cohort using the bisulfite sequencing PCR (BSP-HiSeq). The cohort consisted of 35 stage IA breast cancer patients and 20 healthy volunteers from Nanfang Hospital of Southern Medical University. The results showed that the mean methylation value of each CpG was lower in the cancer patients than in the controls. The optimal cut-off of CpG methylation for breast cancer diagnosis was determined using curves in 35 stage IA breast cancer and 20 healthy samples. This cut-off showed high sensitivity and specificity for breast cancer diagnosis (69.4–83.7% and 85.7–88.6%, respectively) (Figure 6).
Discussion
DNA methylation is a major epigenetic modification that plays a critical role in tumorigenesis. The methylation site tends to be highly modified in CpG islands, which are located within gene promoters or gene bodies, resulting in the activation or inactivation of gene expression. In this study, we analysed the differentially methylated sites of 791 breast tumour samples and 96 normal breast tissue samples from TCGA database and found 23,088 DMCs, with most of the hypomethylated DMCs were located in the CpG islands. This finding was consistent with previous reports, indicating that hypermethylation of CpG island may be a hallmark of breast cancer, may regulate adjacent gene expression, and may be involved in breast tumorigenesis (20-24). We also found that hypomethylation-associated genes were enriched in the olfactory transduction pathway, which confirmed that stimulation of olfactory receptors could promote cancer cell invasiveness and metastasis (25). As reported in previous cancer studies, hypermethylation-associated genes were enriched in five pathways, suggesting the commonality of the cancer epigenome.
In addition, we performed target capture bisulfite sequencing of cfDNA from 10 breast cancer patients and 10 healthy individuals to identify biomarkers that could be used in breast cancer screening. A total of 1,453 cf-DMCs were identified between the breast cancer patients and healthy individuals. As we did in our study, we screened out the CpG sites with abnormal methylation in breast cancer patients through TCGA database and second-generation sequencing, and then annotated the gene pathway and function with KEGG and DAVID respectively, so as to narrow the scope of screening. Unsupervised hierarchical clustering analysis of the methylation status of cf-DMCs could cluster samples of the same group together, even though those 10 patients were at different stages of breast cancer, including stage 0 (ductal carcinoma in situ, DCIS) (n=3), stage 1 (n=3), and stage 2 (n=4). Faryna et al. reported that aberrant methylation occurred in the stage of carcinoma in situ (26). Also, changes in cfDNA can be detected even before neoplasms invade and acquire malignant potential (27). It is reasonable to believe that aberrant methylation in cfDNA can be detected in patients with DCIS, and our research showed that the methylation levels of these CpG sites could differ between patients with early breast cancer and healthy women. Estrogen receptor (ER) positive is the main basis for endocrine therapy in breast cancer patients, but some ER positive breast cancer patients will turn to ER negative in tumor progression. Methylation of ER gene CpG island is one of the mechanisms of ER expression loss, and demethylation will also lead to ER gene reexpression. Moreover, DNA methylation also impact breast cancer subtypes in a certain degree. However, due to the limited number of cases included in our study, there is no correlation analysis in this regard.
A total of 23,088 differential sites were identified between normal and breast cancer tissues, and 14,003 differential sites were identified between breast cancer tissues and WBCs. Furthermore, both T-DMCs and WBC-DMCs showed similar DNA methylation levels in both leukocytes and normal breast tissues, suggesting that the DNA methylation feature of leukocytes is very similar to that of normal breast tissues. However, the number of T-DMCs was significantly greater than that of WBC-DMCs, which might be due to the significantly smaller leukocyte sample size compared to that of normal breast tissue samples. It is possible that the methylation level could be between different samples in the same tissue. To identify the methylation sites in cfDNA for breast cancer diagnosis, we must make sure the methylation difference between patients and healthy controls, and remove the interference of peripheral blood leukocyte DNA, since cfDNA contains leukocyte DNA (to some degree), and they should have consistent changes with breast tumour tissue. Therefore, the aberrant methylation of breast tumours from cancer patients was compared with the methylations in normal tissue, WBCs, and cfDNA from healthy controls.
Thirteen CpG sites (cg04066177, cg04281344, cg05995576, cg06221609, cg08642731, cg11388802, cg12665414, cg14557216, cg19404723, cg19457909, cg24570211, cg25818763, and cg26215982) were identified as diagnostic biomarkers for breast cancer screening. The CpG sites that could serve as biomarkers were annotated to 10 protein-coding genes (FABP9, LCE1A, CACNA1E, CPEB4, DNAI1, DLGAP2, BCAS3, CTNNA2, DCC, and GALNT8). FABP9 and CPEB4 were reported to have high expressions in prostate cancer and breast carcinoma, respectively (28,29). BCAS3 was suggested to have a role in human embryogenesis and tumour angiogenesis (30). CTNNA2 is a tumour suppressor associated with cell-cell adhesion (31). DCC was reported to be hypomethylated in advanced gastric cancer (32). To further verify whether these CpGs could serve as diagnostic markers, we validated three CpGs (cg04281344, cg24570211, and cg26215982) as biomarkers in a new cohort, which showed excellent sensitivity and specificity, suggesting that these CpGs could be used as new diagnostic biomarkers.
The above results demonstrated that DNA methylation is a novel biomarker for cancer diagnosis. However, reliable methods to identify clinically valuable methylated loci remain unclear, as most studies were based on the differences between cancerous and normal tissues (33-35). We comprehensively compared the methylations of tissue and peripheral blood in the context of breast cancer. As a result, we identified, for the first time, 13 CpG sites that could be used as diagnostic biomarkers for breast cancer screening, and validated a new approach to screen methylated biomarkers for cancer diagnosis.
In this study, we developed a method to identify diagnostic methylated biomarkers for breast cancer by comparing the methylations of tissue, WBC, and plasma cell-free DNA. We identified 13 CpGs that may be novel biomarkers for breast cancer diagnosis.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-1128
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-1128). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Esteller M. Epigenetics in cancer. N Engl J Med 2008;358:1148-59. [Crossref] [PubMed]
- Warton K, Samimi G. Methylation of cell-free circulating DNA in the diagnosis of cancer. Front Mol Biosci 2015;2:13. [Crossref] [PubMed]
- Cho YH, McCullough LE, Gammon MD, et al. Promoter Hypermethylation in White Blood Cell DNA and Breast Cancer Risk. J Cancer 2015;6:819-24. [Crossref] [PubMed]
- Radpour R, Barekati Z, Kohler C, et al. Hypermethylation of tumor suppressor genes involved in critical regulatory pathways for developing a blood-based test in breast cancer. PLoS One 2011;6:e16080 [Crossref] [PubMed]
- Yamamoto N, Nakayama T, Kajita M, et al. Detection of aberrant promoter methylation of GSTP1, RASSF1A, and RAR2 2 in serum DNA of patients with breast cancer by a newly established one-step methylation-specific PCR assay. Breast Cancer Research & Treatment 2012;132:165. [Crossref] [PubMed]
- Kloten V, Becker B, Winner K, et al. Promoter hypermethylation of the tumor-suppressor genes ITIH5, DKK3, and RASSF1A as novel biomarkers for blood-based breast cancer screening. Breast Cancer Res 2013;15:R4. [Crossref] [PubMed]
- Swellam M, Abdelmaksoud M, Sayed Mahmoud M, et al. Aberrant methylation of APC and RARβ2 genes in breast cancer patients. IUBMB Life 2015;67:61-8. [Crossref] [PubMed]
- Hao X, Luo H, Krawczyk M, et al. DNA methylation markers for diagnosis and prognosis of common cancers. Proc Natl Acad Sci U S A 2017;114:7414-9. [Crossref] [PubMed]
- Walker DL, Bhagwate AV, Baheti S, et al. DNA methylation profiling: comparison of genome-wide sequencing methods and the Infinium Human Methylation 450 Bead Chip. Epigenomics 2015;7:1287-302. [Crossref] [PubMed]
- Li Q, Suzuki M, Wendt J, et al. Post-conversion targeted capture of modified cytosines in mammalian and plant genomes. Nucleic Acids Res 2015;43:e81 [Crossref] [PubMed]
- Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics 2009;10:232. [Crossref] [PubMed]
- Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009;462:315-22. [Crossref] [PubMed]
- Lister R, Pelizzola M, Kida YS, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 2011;471:68-73. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Avraham A, Cho SS, Uhlmann R, et al. Tissue specific DNA methylation in normal human breast epithelium and in breast cancer. PLoS One 2014;9:e91805 [Crossref] [PubMed]
- Fleischer T, Frigessi A, Johnson KC, et al. Genome-wide DNA methylation profiles in progression to in situ and invasive carcinoma of the breast with impact on gene transcription and prognosis. Genome Biol 2014;15:435. [PubMed]
- Li Z, Guo X, Wu Y, et al. Methylation profiling of 48 candidate genes in tumor and matched normal tissues from breast cancer patients. Breast Cancer Res Treat 2015;149:767-79. [Crossref] [PubMed]
- Lin IH, Chen DT, Chang YF, et al. Hierarchical clustering of breast cancer methylomes revealed differentially methylated and expressed breast cancer genes. PLoS One 2015;10:e0118453 [Crossref] [PubMed]
- Tan J, Gu Y, Zhang X, et al. Hypermethylation of CpG islands is more prevalent than hypomethylation across the entire genome in breast carcinogenesis. Clin Exp Med 2013;13:1-9. [Crossref] [PubMed]
- Rao X, Evans J, Chae H, et al. CpG island shore methylation regulates caveolin-1 expression in breast cancer. Oncogene 2013;32:4519-28. [Crossref] [PubMed]
- Sproul D, Kitchen RR, Nestor CE, et al. Tissue of origin determines cancer-associated CpG island promoter hypermethylation patterns. Genome Biol 2012;13:R84. [Crossref] [PubMed]
- Roessler J, Ammerpohl O, Gutwein J, et al. The CpG island methylator phenotype in breast cancer is associated with the lobular subtype. Epigenomics 2015;7:187-99. [Crossref] [PubMed]
- Sproul D, Nestor C, Culley J, et al. Transcriptionally repressed genes become aberrantly methylated and distinguish tumors of different lineages in breast cancer. Proc Natl Acad Sci U S A 2011;108:4364-9. [Crossref] [PubMed]
- Sanz G, Leray I, Dewaele A, et al. Promotion of cancer cell invasiveness and metastasis emergence caused by olfactory receptor stimulation. PLoS One 2014;9:e85110 [Crossref] [PubMed]
- Faryna M, Konermann C, Aulmann S, et al. Genome-wide methylation screen in low-grade breast cancer identifies novel epigenetically altered genes as potential biomarkers for tumor diagnosis. FASEB J 2012;26:4937-50. [Crossref] [PubMed]
- Izumchenko E, Chang X, Brait M, et al. Targeted sequencing reveals clonal genetic changes in the progression of early lung neoplasms and paired circulating DNA. Nat Commun 2015;6:8258. [Crossref] [PubMed]
- Al Fayi MS, Gou X, Forootan SS, et al. The increased expression of fatty acid-binding protein 9 in prostate cancer and its prognostic significance. Oncotarget 2016;7:82783-97. [Crossref] [PubMed]
- Sun HT, Wen X, Han T, et al. Expression of CPEB4 in invasive ductal breast carcinoma and its prognostic significance. Onco Targets Ther 2015;8:3499-506. [PubMed]
- Siva K, Venu P, Mahadevan A, et al. Human BCAS3 expression in embryonic stem cells and vascular precursors suggests a role in human embryogenesis and tumor angiogenesis. PLoS One 2007;2:e1202 [Crossref] [PubMed]
- Fanjul-Fernandez M, Quesada V, Cabanillas R, et al. Cell-cell adhesion genes CTNNA2 and CTNNA3 are tumour suppressors frequently mutated in laryngeal carcinomas. Nat Commun 2013;4:2531. [Crossref] [PubMed]
- Hibi K, Sakata M, Sakuraba K, et al. Methylation of the DCC gene is lost in advanced gastric cancer. Anticancer Res 2010;30:107-9. [PubMed]
- Johnson K, Houseman E, King J, et al. Normal breast tissue DNA methylation differences at regulatory elements are associated with the cancer risk factor age. Breast Cancer Res 2017;19:81. [Crossref] [PubMed]
- Zouridis H, Deng N, Ivanova T, et al. Methylation subtypes and large-scale epigenetic alterations in gastric cancer. Sci Transl Med 2012;4:156ra140 [Crossref] [PubMed]
- Naumov VA, Generozov EV, Zaharjevskaya NB, et al. Genome-scale analysis of DNA methylation in colorectal cancer using Infinium HumanMethylation450 BeadChips. Epigenetics 2013;8:921-34. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


